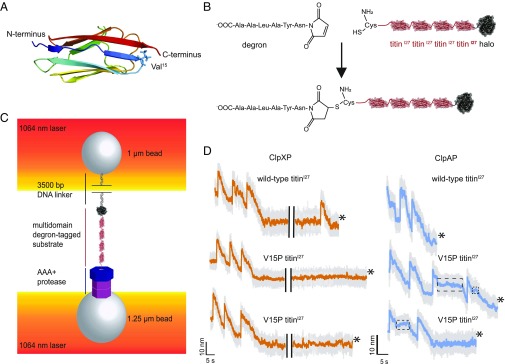
Fig. 1.
Single-molecule N-to-C protein degradation by ClpXP and ClpAP. (A) Structure of the titinI27 domain (Protein Data Bank ID code 1TIT) (cartoon representation), with rainbow coloring from the N terminus (blue) to the C terminus (red). The position of Val15, which is mutated to Pro in the V15P variant, is shown in stick representation. (B) An ssrA-degron peptide was cross-linked to the N-terminal residue of Cys-(titinI27)4-Halo via maleimide-thiol chemistry to generate a substrate for N-to-C degradation. (C) Biotinylated DNA-linked multidomain substrate and ClpAP or ClpXP were attached to separate laser-trapped streptavidin-coated beads in a “dumbbell” configuration for single-molecule optical-trapping experiments. (D) Example traces of single-molecule N-to-C degradation of the multidomain substrate shown in the lower part of B by ClpXP or ClpAP. The bead-to-bead distance changes as the AAA+ protease unfolds (rapid increases) and translocates (gradual decreases) individual substrate domains. The boxed regions of ClpAP traces show pauses during translocation. Traces shown depict raw (gray, 3,000 Hz) and 10 points moving average data (orange for ClpXP and cyan for ClpAP, 300 Hz) under ∼13–16 pN of applied force. In the ClpXP traces, the black bars present a 20-s gap. Asterisks signify tether breaks.