Fig. 4. Truncation of the gRNA traps the HNH domain in the checkpoint intermediate with fewer mismatches on the DNA.
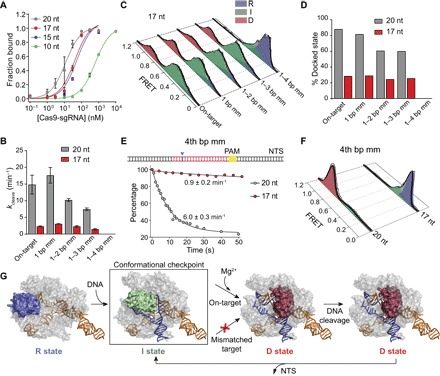
(A) On-target DNA binding assay with 20-nt and truncated gRNAs. (B) Bulk cleavage rates of the DNA substrates by Cas9 assembled with 20- and 17-nt gRNAs. (C) Steady-state smFRET histograms of Cas9 guided with a 17-nt gRNA. (D) D population of Cas9 guided with 20- and 17-nt gRNAs. (E) Top: A single mismatch was introduced at the 4th bp after the PAM-distal end (blue arrowhead). Bottom: Real-time cleavage of 4th bp mm by Cas9 guided with 20- and 17-nt gRNAs. Black curves represent fit to a single exponential decay (mean ± 95% confidence interval). (F) Steady-state smFRET histograms of Cas9 with 20- and 17-nt gRNAs bound to 4th bp mm. (G) Model for substrate-dependent HNH activation. DNA binding triggers transition from R (blue) to I (green) conformation, which serves as a conformational checkpoint between DNA binding and cleavage. In Mg2+, recognition of an on-target locks HNH in the catalytically active D conformation (red), which is destabilized after NTS release. HNH activation is prohibited when the RNA-DNA complementarity drops below a threshold (red cross).
