Figure 2.
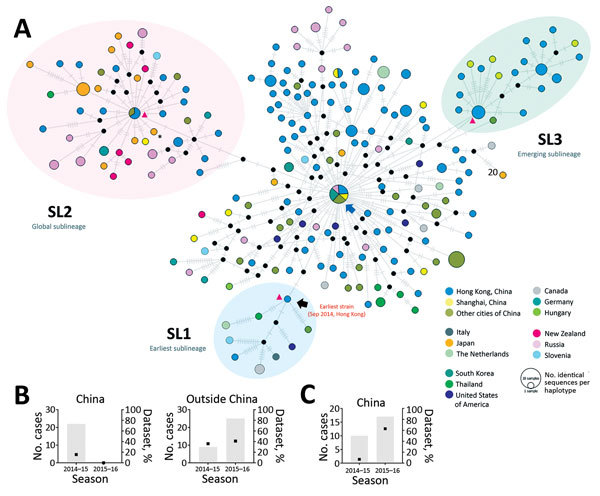
Median-joining haplotype network of 254 complete viral protein 1 nucleotide sequences of norovirus GII.17 Kawasaki. Each vertex represents a unique sampled haplotype. Internal black nodes are unsampled intermediate hypothetical haplotypes. A) Black arrow denotes the first case of norovirus GII.17 Kawasaki in this study (NS-405; collected in September 2014 from Hong Kong). Blue arrow denotes a highly connected basal haplotype from which nearly all haplotypes originated. Vertices are colored by country of collection. Blue shading indicates a sublineage (SL1) genetically closest to the first case of GII.17 Kawasaki virus in this study. Pink shading indicates a sublineage (SL2) with global spread. Green shading indicates an emergent sublineage (SL3) in China in 2016. Vertex size is proportional to the number of sampled sequences sharing the same haplotype. Length of edge is not drawn to scale. Each hatch mark indicates 1 nt difference between connecting haplotypes/nodes. Red triangles represent reference strains of corresponding sublineage (Technical Appendix Table 1). The asterisk denotes the reference sequence of GII.17 Kawasaki virus (Hu/GII/JP/2015/GII.P17_GII.17/Kawasaki308; GenBank accession no. LC037415). Bar charts show the number (gray bars) and percentage (black squares) of cases of sublineages SL2 and SL3 by country in the seasons of 2014–15 (September 2014–June 2015) and 2015–16 (July 2015–March 2016).
