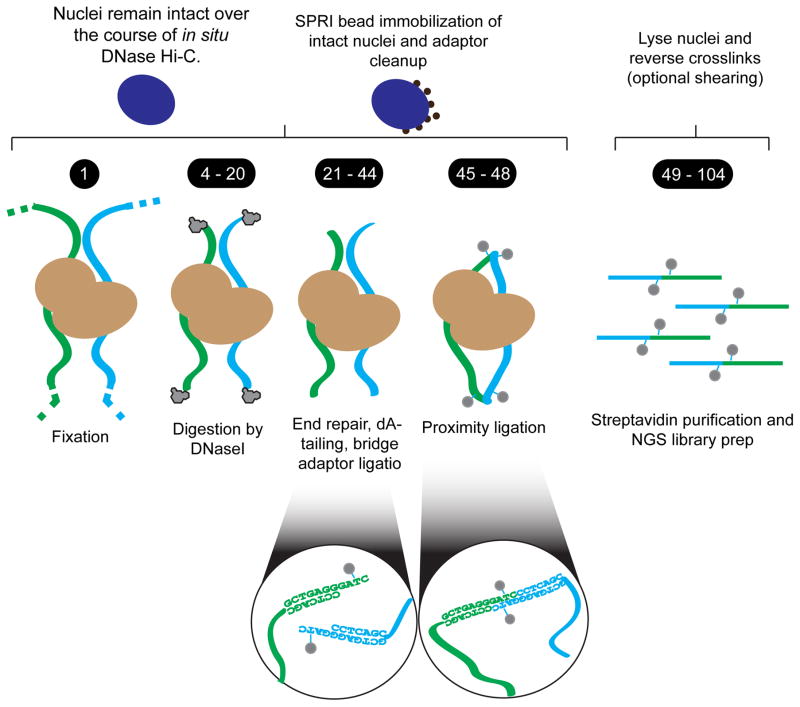
Figure 1. A schematic overview of in situ DNase Hi-C.
First, fixed cells are lysed and digested with the endonuclease DNase I in the presence of divalent manganese—yielding double stranded breaks. Nuclei are then immobilized on carboxylated paramagnetic beads (i.e. ‘AMPure’ beads) to purify intact nuclei and remove free digested DNA fragments. Chromatin is then end-repaired and dA-tailed in situ, and a biotinylated ‘bridge adaptor’ containing a half BamHI site is ligated onto free chromatin ends. Nuclei are then subjected to phosphorylation and in situ proximity ligation, after which DNA is purified and fragments containing ligation junctions are enriched for via streptavidin beads and on-bead Illumina library prep (optionally following sonication).