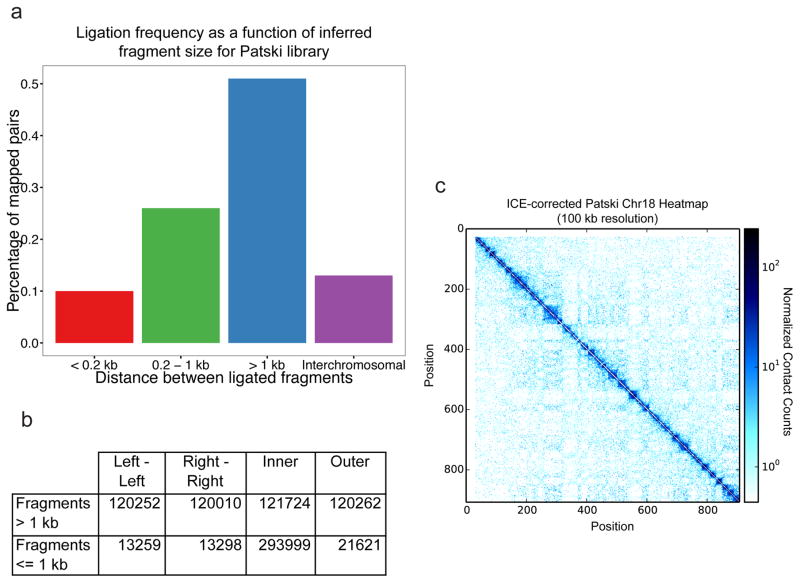
Figure 4. In situ DNase Hi-C results for the mouse embryonic kidney Patski cell line.
a.) In situ DNase Hi-C reads (950,206 downsampled reads from data published in Deng, Ma et al51 (using the mouse Patski cell line, rather than GM12878) demonstrate an enrichment for long-range (i.e. > 1 kb) intrachromosomal read pairs expected of Hi-C libraries. b.) Expected breakdown of mate orientations for read pairs in in situ DNase Hi-C data. For intrafragment distances > 1 kb, a roughly 25% split should be observed for each orientation class. c.) Normalized heat map generated from data published in Deng, Ma et al (GEO Accession: GSE68992) for mouse chromosome 18 at 100 kb resolution. The dataset used to generate this heatmap contained 60,666,200 uniquely mapped, high-quality read pairs.