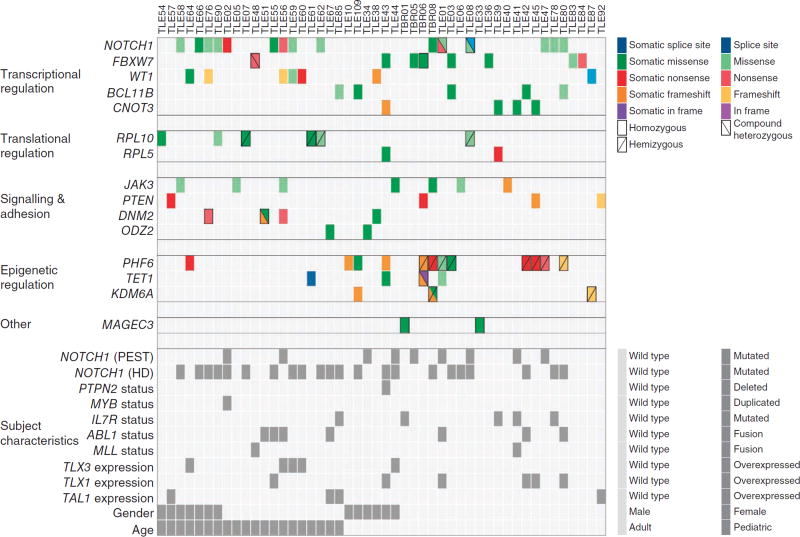
Figure 2.
Overview of mutations in 15 identified candidate T-ALL driver genes in 67 samples from affected individuals. Top, mutations in 15 candidate T-ALL driver genes are shown across the set of cases. For clarity, only affected individuals harboring mutations in at least 1 of the 15 genes are included. Each type of mutation is indicated by color, and symbols indicate whether the mutation was homozygous, hemizygous or compound heterozygous. Mutations with no indication are heterozygous. All mutations shown here were validated by Sanger sequencing. Bottom, the characteristics of the relevant individuals (identified by Sanger sequencing, karyotyping or gene expression analysis) are shown. Mutations in NOTCH1 were hard to identify by exome sequencing owing to low capture efficiency and resulting low sequence coverage of NOTCH1. Detailed descriptions of the mutations shown in this figure are provided in Supplementary Tables 5 and 7–9.