Figure 2. L1 structure and L1-seq validation scheme.
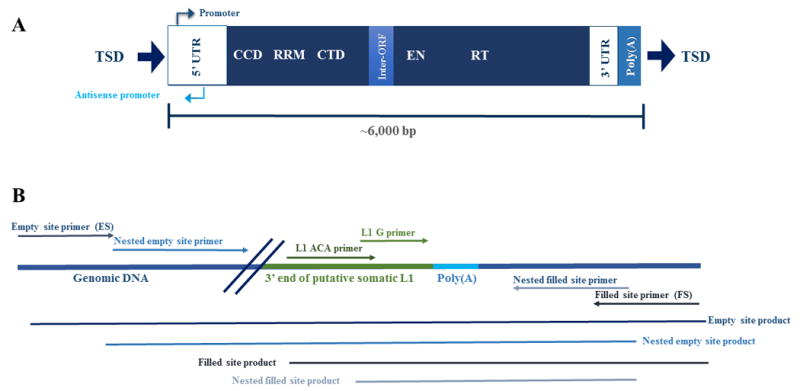
A) LINE-1 structure of a human specific active element in the human genome. Full-length L1 elements are approximately 6,000 nucleotides in length and have a 5’ untranslated region, containing both a promoter and an anti-sense promoter. L1 encodes two open reading frames for proteins on which it relies for its mobilization in the cell (Feng, et al., 1996; Mathias, et al., 1991; Moran, et al., 1996; Scott, et al., 1987). The first open reading frame encodes ORF1p, a protein with RNA chaperoning activity that includes a coiled-coil domain (CCD), an RNA recognition motif (RRM), and a carboxy-terminal domain (CTD) (Martin, 2010; Martin and Bushman, 2001). The second open reading encodes ORF2p, possesses an endonuclease domain (EN) and a reverse transcriptase domain (RT) (Fanning, 1987; Feng, et al., 1996; Mathias, et al., 1991; Moran, et al., 1996). The element also has a poly(A) tail between the 3’ untranslated region and the target site duplication (TSD). Both the poly(A) tail and TSDs are hallmarks of the target-primed reverse transcription, the process by which L1 elements mobilize (Ostertag and Kazazian, 2001). B) The PCR validation method for predicted somatic insertions utilizes two unique areas of sequence in the 3’ end of the L1 element. The sequence ‘ACA’ is incorporated into the L1 ACA primer and is positioned about 90 nucleotides from the poly(A) tail. The ‘ACA’ nucleotides (Boissinot, et al., 2000; Skowronski, et al., 1988) distinguish the human specific transcriptionally active L1 element from other L1s in the genome and allow for a specific PCR product to amplify. The L1 G primer has a ‘G’ nucleotide which is also unique to the human specific active L1 element and the nucleotide is approximately ten nucleotides preceding the poly(A) tail (Ewing and Kazazian, 2010). To validate an insertion, pictured adjacent to a poly-A tail, the L1 primers are used in conjunction with filled and empty site primers to amplify the 3’ end of the somatic insertion. The nested empty site and nested filled site primers are flanked by the empty and filled site primers and are used to verify that insertions predicted in tumor are truly absent from normal tissue. Additionally, nested primers are utilized when amplifying low copy number somatic insertions because they are able to detect when one cell out of a thousand has a copy of an insertion. The size of the expected products and relative locations of the primers are pictured in the figure.
