Figure 1.
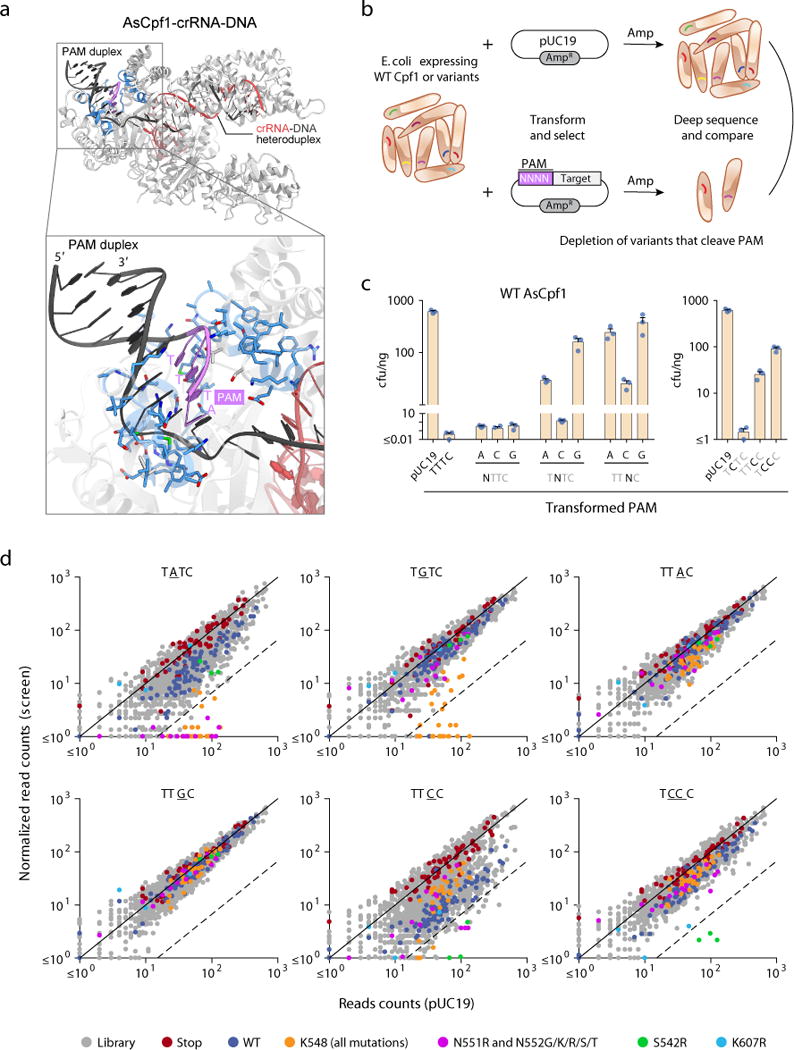
A bacterial interference-based negative selection screen identifies amino acid substitutions of AsCpf1 conferring activity at non-canonical PAMs. (a) Crystal structure of AsCpf1 (PDB ID: 5B43) in complex with crRNA and target DNA, highlighting the PAM nucleotides (magenta), and PAM-proximal residues selected for mutagenesis (blue). (b) Schematic of bacterial interference assay used to identify variants with altered PAM specificity. (c) Sensitivity of wild-type AsCpf1 to substitution mutations in the PAM as measured by bacterial interference. Bars show mean ± s.e.m. of n = 3 plated transformations. (d) Scatter plots of screen readout, highlighting depleted variants. Each dot represents a distinct wild-type or mutant codon. The dashed line indicates 15-fold depletion.
