Figure 1.
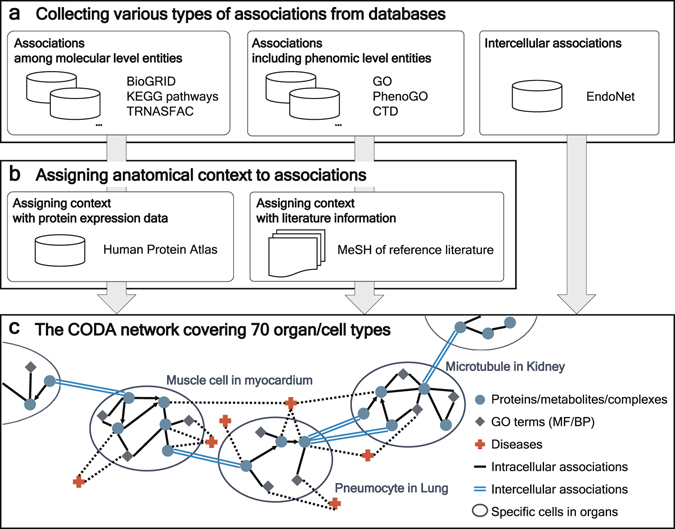
Overview of constructing the CODA network. (a) Associations including both molecular level entities and phenomic level entities are gathered from diverse databases, BioGRID, KEGG pathways, TRANSFAC, GO, PhenoGO, CTD, and EndoNet. All of the associations do not include anatomical contexts at first except for intercellular associations from EndoNet. (b) Anatomical contexts are assigned to associations among molecular level entities by using protein expression data from HPA. For associations including phenomic level entities, anatomical contexts are added to the associations using MeSH of their reference literature. Intercellular associations have anatomical context ab initio. (c) As a result, constructed CODA network consists of not only organ/cell type specific networks but also intercellular associations. Diverse associations among molecular level entities and phenomic level entities with anatomical context are contained in CODA network.
