Figure 1.
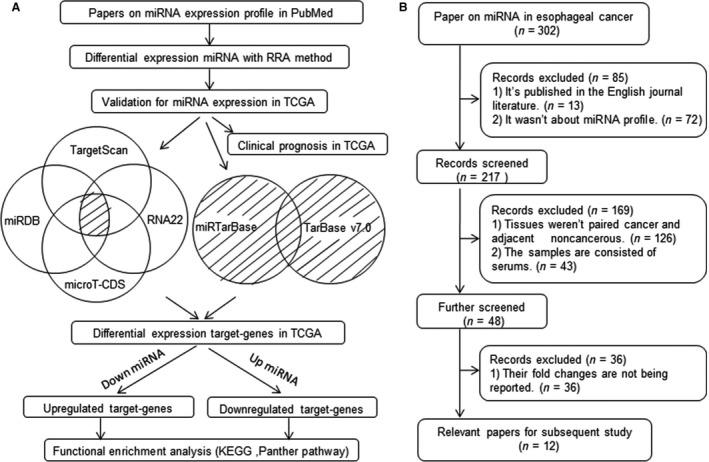
Flowchart for the study. (A) General strategy of the study. The RRA method (novel robust rank aggregation) was applied to obtaining significant miRNAs. Six databases including TargetScan, miRDB, microT‐CDS, RNA22, miRTarBase, and TarBase v7.0 were applied to obtaining the target genes of the miRNA. The dashed area in A represents the target genes we need. TCGA: The Cancer Genome Atlas. (B) Screening rules for articles on the miRNA expression profile in PubMed.
