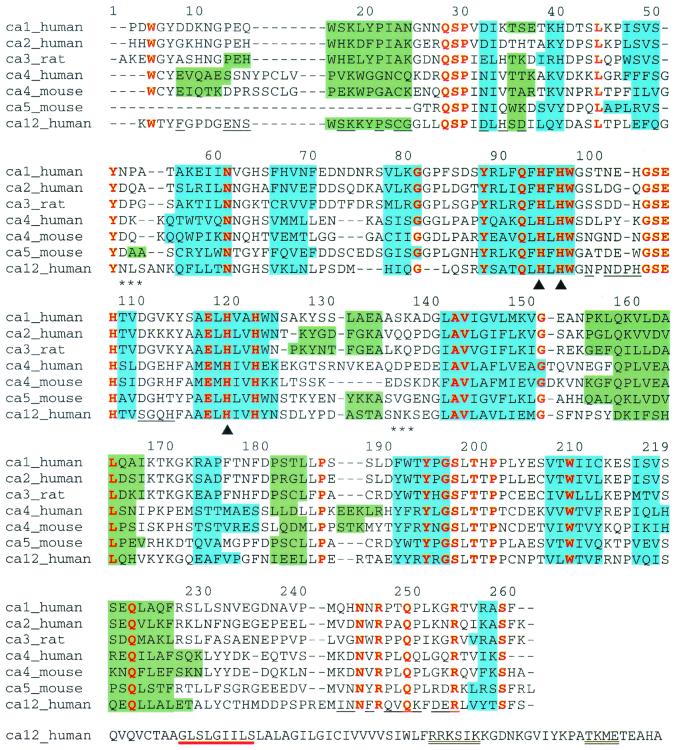
Figure 1.
Sequence alignment of human CAs I (RCSB accession code 1CAB; ref. 61), II (1CA2; ref. 47), IV (1ZNC; ref. 35), and XII, mouse CAs IV (2ZNC; ref. 54) and V (1DMY; ref. 58), and rat CA III (1FLJ; ref. 62) based on the alignment of secondary structural elements as observed in the individual crystal structures. Blue and green shading denote β-strands and α-helices, respectively. Residues conserved throughout all of these sequences are colored red and residues at the CA XII dimer interface are underlined. Triangles (▴) mark histidine–zinc ligands and asterisks (*) denote asparagine glycosylation sites. The alignment is numbered according to the sequence of human CA II. The entire sequence of CA XII is shown, and the GXXXG and GXXXS motifs that signal transmembrane helix dimerization are underlined in red; potential phosphorylation sites are double underlined. For all other isozymes, the N and C termini are truncated according to what is visible in each crystal structure.