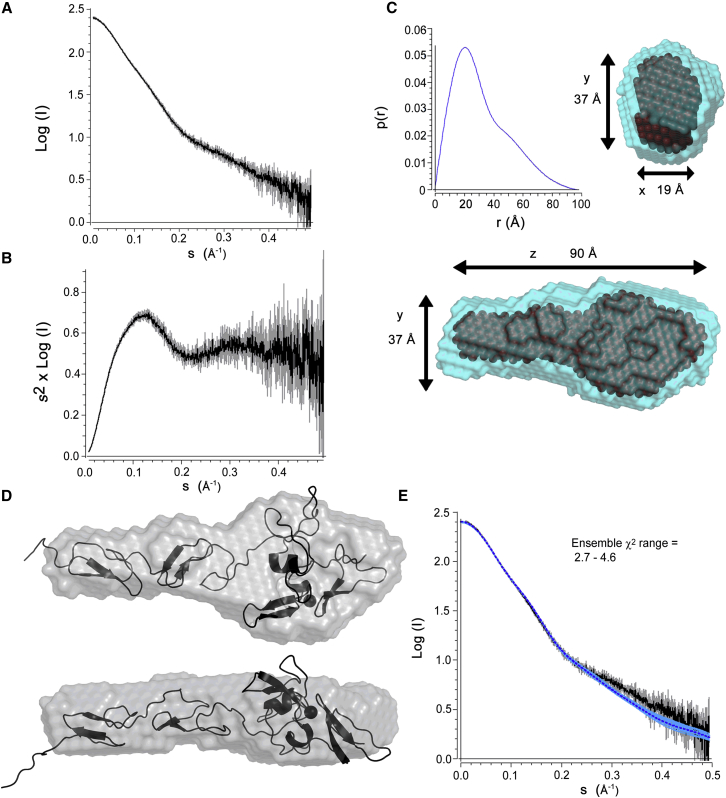
Figure 4.
SAXS Data for FBN1E2cbEGF1 Reveals a Linear Shape
(A) Scaled, merged, and averaged X-ray scattering curves collected with purified FBN1E2cbEGF1 at concentrations of 11.55, 5.78, 2.89, and 1.44 mg/mL. Analysis of these data confirm that the protein behaves as a monomer in solution.
(B) Kratky plot of scaled, merged, and averaged SAXS data showing a peak falling to a plateau; this behavior is characteristic of a folded and relatively rigid protein.
(C) P(r) distribution and ab initio modeling of particle shape using DAMMIF. The blue transparent surface represents the shape produced from averaging the 20 independently generated ab initio structures with DAMAVER, and the darker spheres within this envelope represent the core shared particle shape calculated by the DAMFILT algorithm. Fitting this DAMFILT particle shape to the scattering data gave a χ2 value of 0.7240.
(D) Comparison of a selected model (black cartoon) from the NMR structural ensemble with the envelope produced by DAMFILT ab initio modeling (gray surface).
(E) Fitting of the 20 structures in the NMR ensemble to the SAXS data. The SAXS data and error bars are shown in black and gray, respectively. The fits of the NMR structures are shown in blue with a dark blue dashed line showing the fit of model 1 of the NMR ensemble.
The error bars in (A), (B), and (E) are derived from the SCATTER software package using the data collected at four proteins concentrations.