FIG 2 .
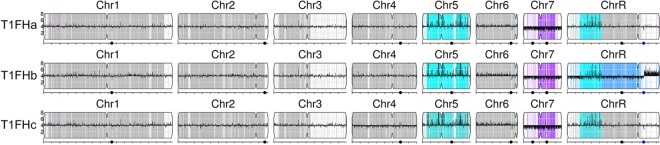
YMAP visualization of SNP-CGH data for three T1FH clones. The copy numbers and SNP-allele ratios of all three T1FH samples were visualized with YMAP (49). Changes in the copy number estimate for regions relative to the parental strain are illustrated by dark bars drawn up- or downward, depending on the direction and magnitude of the change. These strains appear to be tetraploid on the basis of the copy number estimates for Chr7 (and most of ChrR in clone T1FHb), which are 3/4 of the other chromosomes. Color illustrates SNP status across regions. Heterozygous regions are gray, white regions do not have SNPs in the SC5314 reference sequence, and cyan is homozygous “a” alleles (e.g., aaaa on Chr5 in all three clones and ChrR in T1FHa and T1FHc and aaa in T1FHb). Intermediate ratios are indicated by intermediate colors. Blue shade represents more copies of “a” alleles (e.g., aab on the central trisomic region of ChrR in T1FHb), and purple shade represents more copies of the “b” alleles (e.g., abb on Chr7 in all three clones). The portion of ChrR in clone T1FHb to the right of the rDNA region (blue dot) is present in five copies. Major repeat sequences are represented by black dots.