Chronic lymphocytic leukemia (CLL) is a slowly developing progression-prone disease. MicroRNAs miR-155 and miR-150 are small inhibitors of gene expression in B-cells that were previously connected to the pathogenesis of CLL. We herein evaluated relationship of miR-155/miR-150 network with clinical and routine laboratory parameters of the CLL patient cohort utilizing multivariate analyses and found its association with overall survival and progression of CLL.
Aggressive course in CLL affects ~20–40% patients as indicated by the clonal-evolution data.1 One of the strongest aggressive course associates is 17p13 deletion occurring in 5–10% of CLL patients.2 Other risk associates were previously connected with the B-cell receptor signaling, including the CD38, ZAP-70, and intact variable region of IgH (IgHV).3, 4, 5 Recently, miR-155 has been connected with CLL aggressiveness6 and progression of the monoclonal B-cell lymphocytosis (MBL).7 Our group suggested that it is the transcriptional upregulation of the miR-155 host gene (MIR155HG) via the Myeloblastoma proto-oncogene (MYB) that lead to the increased level of the mature miR-155 in CLL, which may represent the molecular pathway of CLL aggressiveness.8 In addition, the downstream events of miR-155 overexpression, including downregulation of transcription factor PU.1 may also coincide with CLL aggressiveness and patient outcome.8, 9, 10
Another miR-155 target is the Src homology-2 domain-containing inositol 5-phosphatase 1 (SHIP1), which supposedly blocks the B-cell receptor signaling (BCR).11 Indeed, treating the CLL or normal B-cells with the CD40-ligand or the B-cell-activating factor upregulated the miR-155 level and enhanced sensitivity to the BCR ligation, effects that could be blocked by inhibitors of miR-155.11 Furthermore, another microRNA involved in the B-cell differentiation, miR-150, has also been shown to interfere with the BCR signaling by additional mechanisms involving its targets GAB1 and FOXP112 and presumably also through its better-recognized target MYB.13 Such possibility was also supported elsewhere.14 Importantly, while the miR-155 and the miR-150 targets (such as MYB) are rather negative CLL predictors, the levels of miR-150 and the miR-155 targets (SHIP1, PU.1) could be considered as the positive predictors, although the complete network has not been tested yet. Both miR-15511 and miR-15012 were validated as the crucial components regulating sensitivity to the BCR ligation in CLL.
We herein investigated how levels of miR-155, miR-150 and their network associate with clinical outcome of 127 CLL patients (36 females and 91 males, age range 36–88; median age 65, median follow-up was 54 months, see Supplementary Table, ST1). These patients donated peripheral blood (PB) samples during years 2009–2013. 74% of all patients are currently alive. 23% of all patients entered the study as previously treated. Fludarabine-Cyclophosphamide-Rituximab (FCR) regimes were applied as the first line of therapy in 69%. Expression levels of microRNAs and their target mRNAs were determined in the purified CD19+ PB cells (Supplementary Material 1). The statistically evaluated variables were: absolute number of PB leukocytes (WBC) and Lymphocytes, %PB monoclonal lymphocytes, platelets, IgHV, CD38, ZAP-70, Richter’s transformation, cytogenetics: del(17)(p13) (TP53), del(11)(q22.3) (ATM), trisomy-12 (t12), del(13)(q14) and finally the levels of the studied miR-155/miR-150 network (PU.1, MYB, miR-150, miR-155, see Supplementary Material 2).
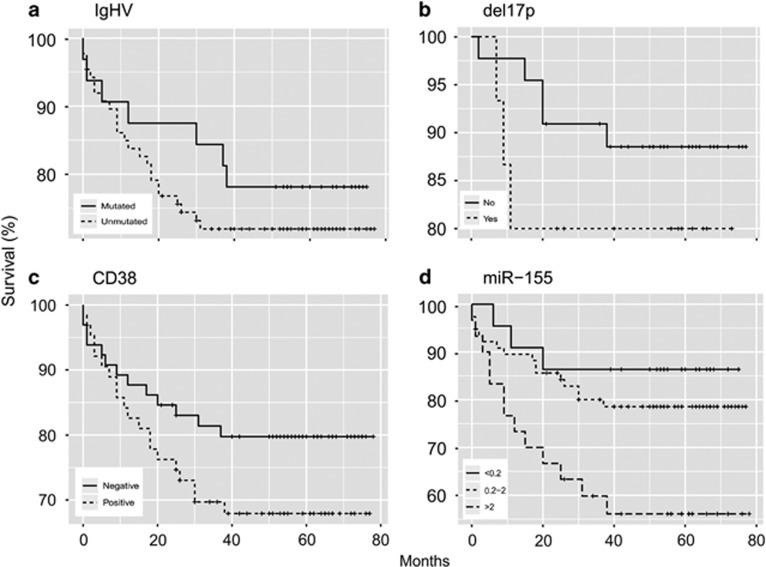
To address our primary goal, we generated the Kaplan–Meier overall survival (OS) plots to document the CLL patient survival in relation to the variables. Figure 1 shows that expectedly the unmutated status of IgHV (a), CD38 expression (c) as well as del17p (b) were detected in patients with lower OS unlike those that lack IgHV mutation, CD38 expression or 17p deletion. The levels of miR-155 were divided into three expression intervals and their associations to OS presented in univariate setting (Figure 1d). The OS reached plateau at 86% for low miR-155 level, 79% for intermediate level and 56% for high-miR-155 level. High-miR-155 subgroup contained ~22% of CLL patients. Next, to achieve the primary goal, we utilized the multivariate Cox proportional hazards model with interactions to delineate role of each member of the miR-155 network (miR-150, miR-155, MYB, PU.1) in comparisons to standard variables, including IgHV, CD38, del17p and others (for detail see Supplementary Figure 1 and extended statistical description in the Supplementary Material 3). To test the interlinked effects of the variables, one must evaluate miR-155/miR-150 network in patients that are CD38-positive versus those that are CD38 negative as the CD38-positive patients are in general at 170.7 times higher risk that the CD38-negative patients (P-value 0.0110). Similarly, the variables were also tested separately whether the patient contains 17p deletion or not; as the presence of del(17)(p13) causes 89.31 times higher risk of dying (P-value 0.0086). MiR-155 and its upstream positive regulator, the oncogenic transcription factor MYB, represent according to our multivariate testing, an important negative risk factor of the CLL patient fate (P=0.0278 and P=0.0438, respectively, Supplementary Material 3). In contrast, the levels of miR-150 that inhibits MYB associated with the prolonged OS (P=0.0307) in CLL (Supplementary Material 3). The levels of PU.1 that is a validated target of the miR-155 were at borderline significance, for details see Supplementary Material 3. To conclude this part, as indicated by univariate testing and further evaluated by multivariate tests, the miR-155 network appears to significantly influence the CLL patient fate, leading to the conclusion that high levels of oncogenic miR-155 and its regulator oncogene MYB are the variables interacting statistically with the low levels of the tumor suppressors: miR-150 and PU.1. This mechanism is discussed bellow.
Figure 1.
MiR-155 levels are increased in CLL patients with lower OS. Univariate Kaplan–Meier estimators of the overall survival documenting survival (%, Y axis) over time (months, X axis) in relation to the variables: IgHV status (a), del17p status (b), CD38 expression (c) and miR-155 levels (d). Numbers of patients in (a–c) are listed in the ST1; (d) MiR-155 (low: 22, intermediate: 76, high: 28 patients). The positivity or levels for each variable is indicated by lines (dashed or full). The legend is located on the left bottom part of each picture.
To test the miR-150/miR-155 network in a more dynamic manner, we tested its role in respect to the CLL progression through clinically distinct disease phases. These phases reflect the stability of CLL, its progression, and therapy-requirement for both untreated (phases 1–3) as well as previously treated CLL patients (phase 4–6) (see Supplementary Material 4 for details). For example, if the patient was never treated and has a stable clinical behavior, it belongs to the phase 1, which is a phase not requiring any therapy; or if the CLL progresses, it enters phase 2; and finally upon indication of therapy, the patient enters phase 3. Similar phases are developed for the previously treated patients (phase 4–6) and these phases may repeat upon next line/s of the therapy. The samples in phases 4–6 are slightly underrepresented compared to phases 1–3 (31 vs 98 samples). We utilized multivariate testing (proportional odds model as a special case of cumulative logit models, see Supplementary Material 5) and assessed the levels of all variables to affect the belonging to one of the six CLL disease phases. Number of patients in each phase had negligible effect. The table within the Supplementary Material 5 data also contains a list of parameter estimates accompanied by P-values. Out of 19 disease characteristics, only CD38 (P-value 0.033), lymphocytosis (P-value 0.006), Trisomy12 (P-value 0.010), deletion17p (P-value 0.035), low platelets (P-value 0.009) and miR-155 (P-value 0.014) had statistically significant influence on progressing through CLL phases. Supplementary Figure 1 depicts how miR-155 at different expression levels (0.01, 0.1, 1, 2, 5) can increase the probability of a patient to progress into a higher disease phase. This is shown in respect to the other significant variables (CD38, Lymphocytes, Trisomy12, Deletion17p and platelets). To conclude this part, miR-155 is the only variable (P=0.014) within the tested members of the miR-155/miR-150 network that has a significant impact on belonging to a more advanced disease phase. The CD38 positivity (P=0.033), lymphocytosis (P-value 0.006), Trisomy12 (P-value 0.010), Deletion17p (P-value 0.035), lower platelets (P-value 0.009) had similar effects.
We herein provide evidence of the association of miR-155 and its network with the clinical outcome of a CLL cohort of 127 patients with a 54-month follow-up. As expected, the role of some currently known predictive variables (17p deletion and CD38) was confirmed.2 Using the Cox proportional hazards model, we newly associate the level of miR-155/miR-150 network components with a patient’s survival and risk of a patient’s death (Supplementary Figure 2). The evidence that miR-155 represents adverse factor and miR-150 a beneficial factor were previously suggested.8, 11 However, to our best knowledge, it has not been tested together and analyzed by multivariate tools. The importance of miR-150 together with miR-155 strongly supported the current view of the importance of BCR signaling for the CLL pathogenesis and prognostication.12 Confirming the roles of CD38 and del(17)(p13) as negative parameters for the survival, as well as the risk of death provide independent support to our data. Furthermore, providing the role of miR-155 levels in progression through the CLL disease phases (Supplementary Figure 1) together with the data of CD38, lymphocytes, platelets and cytogenetics suggested the fundamental importance of miR-155 for the CLL prognostication.
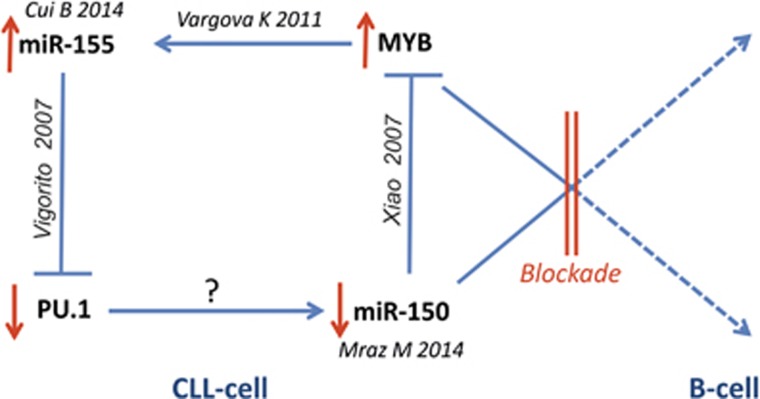
We are proposing an updated model for CLL pathogenesis, which involves the miR-155/miR-150 network. As summarized in the Figure 2, miR-150 downregulation appears to be the initial event during B-cell differentiation, resulting in defective inhibition of MYB.13 Inappropriate levels of MYB transcriptionally upregulate miR-155.8 MiR-155 blocks the expression of PU.1, which is a well-known regulator of differentiation in B-cells.15 A miR-155/PU.1 axis inhibition was previously shown to affect production of high-affinity IgG1 antibodies.15 Finally, the miR-155/miR-150 network’s dysregulation11, 12 may also account for the BCR signaling disruption, which is an integral part of the CLL phenotypic features.
Figure 2.
Model of the pathogenic blockade. Indicated by red lines, the B-cell differentiation and CLL pathogenesis involve miR-155/miR-150 network, including its targets MYB and PU.1. Citations are adjacent to the roles of particular molecules. Activation is indicated by arrow, inhibition is indicated by T-shaped arrow.
Acknowledgments
This work was funded by GAČR 16-05649S, AZV 16-27790A, LH15170, UNCE 204021, Progres Q26, LQ1604 NPU II, and CZ.1.05/1.1.00/02.0109, MP: GAČR P402/12/G097 and IAP research network grant No. P7/06, AB, ZZ, KM: RVO-VFN64165 and Progres Q28.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
Authors contribution
KV molecular biology; MP statistics; JV, LM, ND, PO, MT clinical data; AB, KM, ZZ: cytogenetics; TS: research design.
The authors declare no conflict of interest.
Supplementary Material
References
- Rodriguez-Vicente AE, Diaz MG, Hernandez-Rivas JM. Chronic lymphocytic leukemia: a clinical and molecular heterogenous disease. Cancer Genet 2013; 206: 49–62. [DOI] [PubMed] [Google Scholar]
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 2000; 343: 1910–1916. [DOI] [PubMed] [Google Scholar]
- Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 1999; 94: 1848–1854. [PubMed] [Google Scholar]
- Ibrahim S, Keating M, Do KA, O'Brien S, Huh YO, Jilani I et al. CD38 expression as an important prognostic factor in B-cell chronic lymphocytic leukemia. Blood 2001; 98: 181–186. [DOI] [PubMed] [Google Scholar]
- Crespo M, Villamor N, Gine E, Muntanola A, Colomer D, Marafioti T et al. ZAP-70 expression in normal pro/pre B cells, mature B cells, and in B-cell acute lymphoblastic leukemia. Clin Cancer Res 2006; 12(3 Pt 1): 726–734. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Croce CM. Role of microRNAs in lymphoid biology and disease. Curr Opin Hematol 2011; 18: 266–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrajoli A, Shanafelt TD, Ivan C, Shimizu M, Rabe KG, Nouraee N et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood 2013; 122: 1891–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargova K, Curik N, Burda P, Basova P, Kulvait V, Pospisil V et al. MYB transcriptionally regulates the miR-155 host gene in chronic lymphocytic leukemia. Blood 2011; 117: 3816–3825. [DOI] [PubMed] [Google Scholar]
- Rossi S, Shimizu M, Barbarotto E, Nicoloso MS, Dimitri F, Sampath D et al. microRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood 2010; 116: 945–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visone R, Rassenti LZ, Veronese A, Taccioli C, Costinean S, Aguda BD et al. Karyotype-specific microRNA signature in chronic lymphocytic leukemia. Blood 2009; 114: 3872–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B, Chen L, Zhang S, Mraz M, Fecteau JF, Yu J et al. MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood 2014; 124: 546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mraz M, Chen L, Rassenti LZ, Ghia EM, Li H, Jepsen K et al. miR-150 influences B-cell receptor signaling in chronic lymphocytic leukemia by regulating expression of GAB1 and FOXP1. Blood 2014; 124: 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 2007; 131: 146–159. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang Z, Dai X, Pan J, Ge J, Han X et al. Re-expression of microRNA-150 induces EBV-positive Burkitt lymphoma differentiation by modulating c-Myb in vitro. Cancer Sci 2013; 104: 826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 2007; 27: 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.