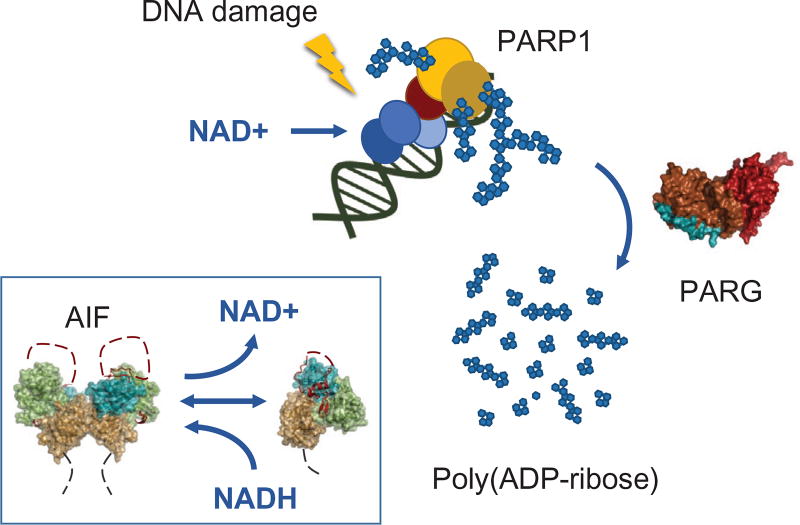
Fig. 1.
NAD as a metabolic cofactor, damage sensor, and signal in the interrelated PARP, PARG, and AIF allosteric responses that control cell responses to DNA damage. PARP1 acts in the cellular response to ssDNA breaks, where it links damage binding to acute production of poly(ADP-ribose) (PAR) that recruits multiple repair factors to the damage site. PARP1 activation by DNA damage entails unfolding of an autoinhibitory domain to enable proper positioning of NAD+ that leads to a burst in PAR production (PARP1 [based upon PDB 4DQY]—ART domain, gold; HD domain, yellow; WGR domain, red; zinc fingers 1–3, blue. PARG—macrodomain fold, red; mitochondrial targeting sequence, cyan). AIF is an allosteric sensor controlling mitochondrial metabolism and programmed cell death based upon NAD levels that control AIF monomer to dimer assembly by local unfolding (FAD domain, green; NADH domain, gold; C-terminal domain, teal; C-loops, red-dotted lines; mitochondrial innermembrane tethers, dotted gray lines).