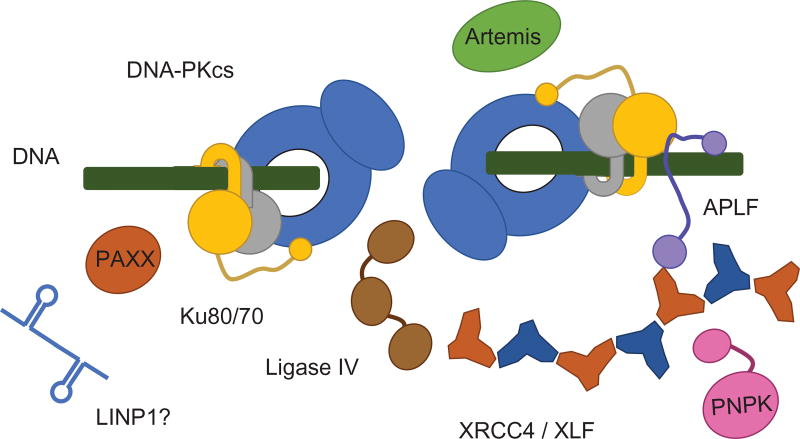
Fig. 3.
NHEJ as a dynamic assembly around Ku dimers at DNA ends. The architecture of the NHEJ core complex based on combined crystallographic and SAXS structures uncovers mechanisms for the synergy of DNA-PKcs/Ku/XRCC4/XLF/LigaseIV interactions in ligating DSBs. Unstructured linkers join DNA-PKcs and Ku dimers in the architectural control of DNA ends by XRCC4–XLF filaments and flexible scaffolding by APLF, PAXX, and possibly RNA (LINP1). The resulting dynamic assembly positions and protects ends while allowing access to Artemis and PNKP for end processing and Ligase IV for end joining (DNA-PKcs, blue; Ku70, gray; Ku80, yellow; XRCC4, orange; XLF, dark blue, PAXX, dark orange; Artemis, green; PNKP, pink; APLF, purple; Ligase IV, brown).