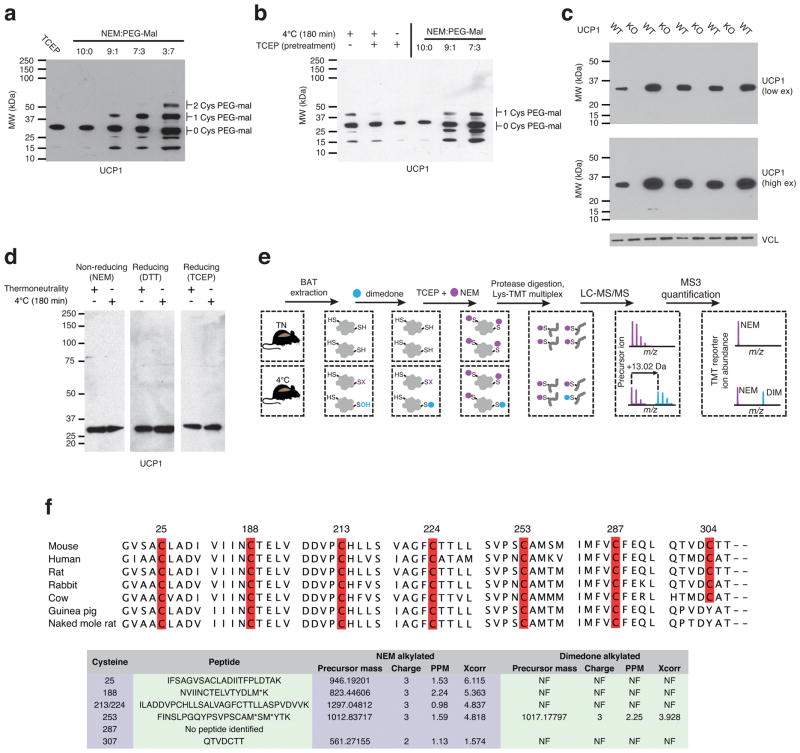
Extended Data Figure 4.
Assessing UCP1 reversible cysteine oxidation status in vivo by immunoblot and mass spectrometry. a, Calibration of UCP1 cysteine gel shift immunoblot. Calibration of cysteine-dependent shifts by incubation of BAT protein with TCEP and different ratios of NEM and PEG-Mal indicates that a single PEG-mal labelling event shifts UCP1 by ~10 kDa above the native molecular weight. b, Calibration of UCP1 cysteine oxidation status indicates that the gel shift observed upon cold exposure (lane 1) is cysteine dependent, as TCEP pretreatment results in a loss of the shift (lane 2). In addition, the cysteine-dependent mass shift is due to a single oxidation event as determined by including the calibrating markers (lanes 4–6). c, Calibration of specificity of UCP1 antibody in BAT. d, Reducing and non-reducing SDS-PAGE analysis of UCP1 to monitor cysteine dependent and independent inter-protein interactions during acute cold exposure. e, Scheme for identification of sulfenylated cysteines on UCP1 by dimedone labeling and mass spectrometry. Following acute cold exposure BAT protein thiols are differentially alkylated with dimedone to selectively label sulfenylated thiols and NEM to label non-sulfenylated thiols. Samples are subjected to trypsin digestion, followed by Lys-TMT labeling, and MS quantification of UCP1 cysteine containing peptides in their dimedone and NEM alkylated forms. Two technical points should be noted in this strategy when interpreting relative quantitation of NEM and dimedone-alkylated peptides. First, these differently alkylated peptides may not necessarily ionize with the same efficiency. Second, NEM is reported to react with sulfenic acids albeit less efficiently than with free thiols32, which should be factored in when considering the order of addition of dimedone/NEM and potential underestimation of sulfenylation status. f, (Top) Amino acid sequence alignment of UCP1 proteins highlighting the candidate cysteine residues contained within the mouse protein and their level of conservation across various species. (Bottom) Summary of MS determination of UCP1 cysteine sulfenylation status. Six of seven UCP1 cysteines were identified, with all but one being identified exclusively in the unmodified (NEM-alkylated state). Cys253 is identified as dimedone labeled, indicating that it is a site for sulfenylation.