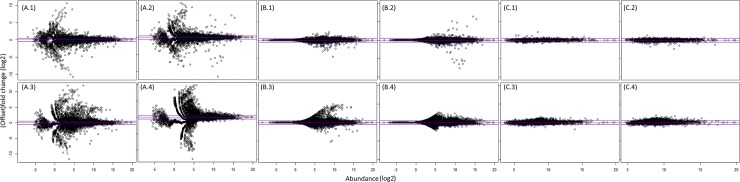
Fig 3. Distribution of DE as calculated by using fold change (FC) versus offset fold change (OFC) and the effect of incorporating hierarchical DE (HDE).
Shown are MA plots (x-axis showing gene abundance (log2), y-axis indicating FC/OFC for replicate-to-replicate comparisons for the 2h samples. Panels A1, B1, C1 show 02-A comparisons, panels A2, B2, C2 02+A samples, A3, B3, C3 for 02-H and A4, B4, C4 for 02+HT samples (sample codes: 02 = 2h of exposure, A = abdomen, HT = head-thorax, + = with rivals,— = without rivals). Panel A shows the distribution of DE calculated using FC, highlighting how the low abundance genes distorted the distribution of DE. Panel B shows the DE distribution using OFC (offset = 20). Here the low abundance genes were no longer underlined as DE. Panel C shows the DE distribution following hierarchical DE analysis using OFC for A- and HT-specific genes emphasizing the effect of excluding potentially leaky genes. The red horizontal lines denote 0 log2 FC/OFC and the blue lines ± 0.5 log2 FC/OFC.