Figure 3.
Transcriptome Analyses of M+ST14hi and M+ST14lo Duct Cells
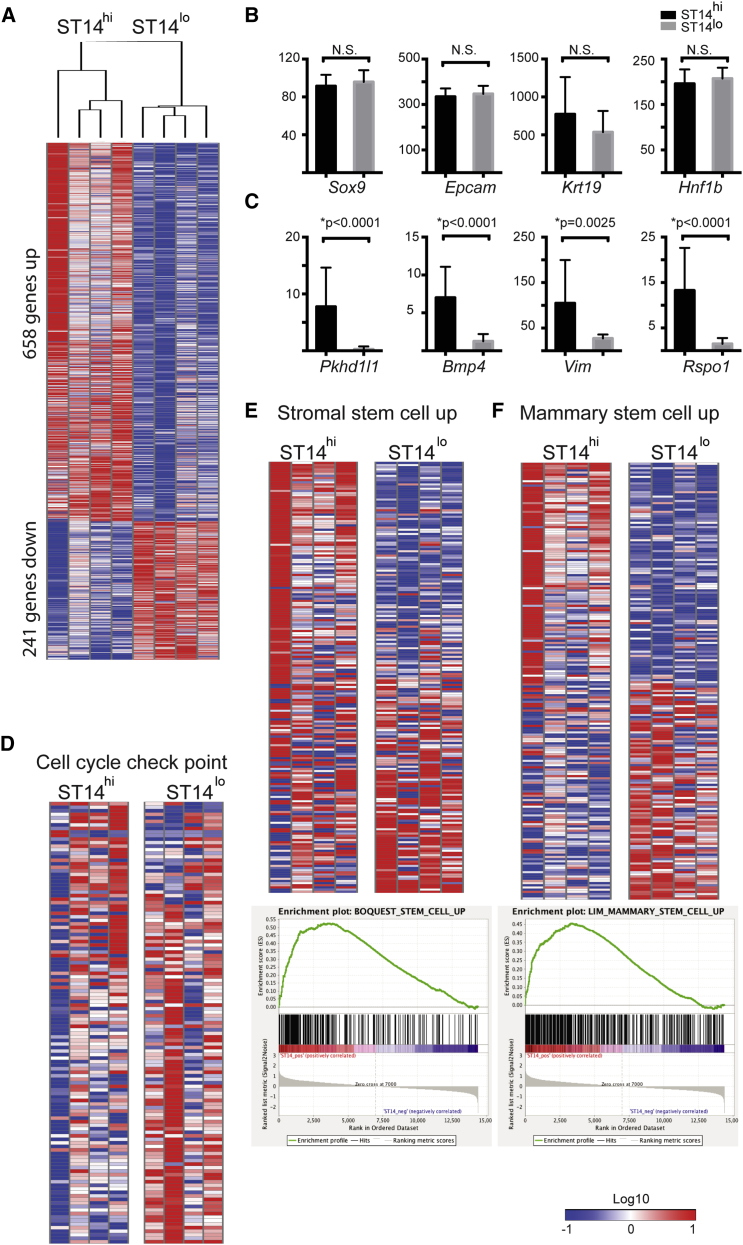
(A) Kendal's tau unsupervised clustering of RNA-seq data of the M+ST14hi and M+ST14lo populations (n = 4 independent experiments for each population).
(B and C) RNA expression levels of selected individual genes. The y axis indicates RPKM (reads per kilobase per million). N.S., not significant. (B) Prototypical cholangiocyte marker expression levels were comparable in the duct populations. (C) Pkhd1l1, Bmp4, Vim, and Rspo1 are examples of differentially expressed genes.
(D–F) Representation of differentially expressed gene set enrichment analysis categories. (D) Downregulated cell-cycle checkpoint genes. (E) Upregulated in stem cells (BOQUEST) (Boquest et al., 2005). (F) Upregulated in mammary stem cells (Lim et al., 2010).