Figure 2.
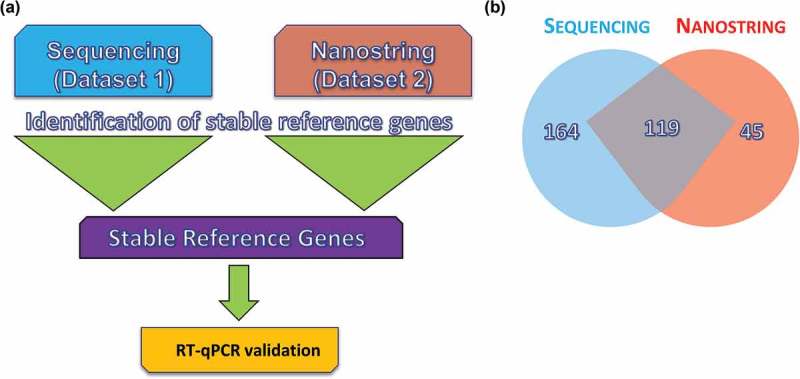
Workflow for the reference gene identification method. Reference genes were identified using small RNA sequencing and a chip-based method. Each data set was unique and included different donors and diverse conditions. Identification of reference genes from each data set was conducted in parallel using the four major algorithms for reference gene identification (NormFinder, GeNorm, BestKeeper and Delta Ct). (a) Common microRNAs (miRs) were selected from each set for further validation using reverse transcription–quantitative polymerase chain reaction (RT-qPCR) in a third unique sample set. (b) Venn diagram showing miRs identified by sequencing compared to those identified by NanoString. Data are representative of the two donors in common between data sets 1 and 2.
