Figure 7.
Pluri-IQ Application Pipeline and Its Performance Evaluation in Immunofluorescence Images
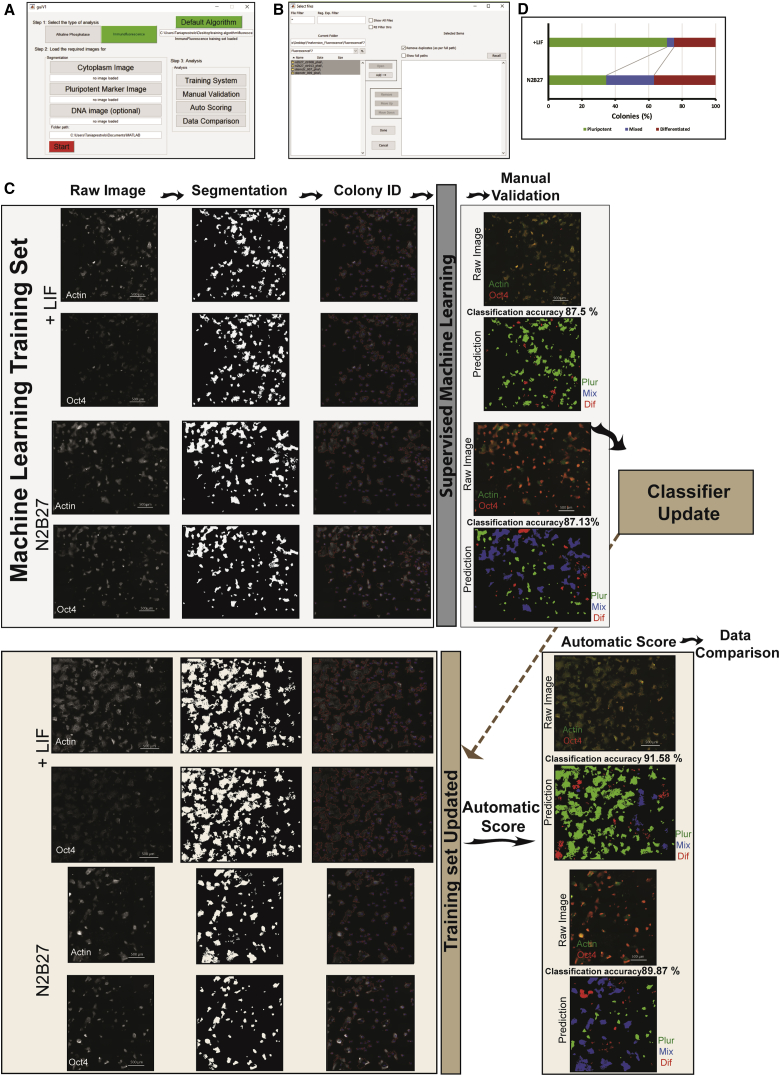
(A) The main graphical user interface (GUI) of Pluri-IQ.
(B) GUI used to select different folders containing the images to perform manual validation, autoscoring or data comparison.
(C) Pluri-IQ pipeline: two different images with different degrees of pluripotency were used to create the machine-learning training set (upper panel). After each channel segmentation and colony identification, a fluorescence training set was created and manually validated. After the classifier automatic update, two new images were scored automatically by Pluri-IQ and classification accuracy was evaluated (bottom panel). Scale bar, 500 μm (in raw images). Color code on the raw images: green, actin; red, OCT4. Color code on the images prediction: green, pluripotent colonies (Plur); blue, mixed colonies (Mix); red, differentiated colonies (Dif).
(D) Percentage of pluripotent, mixed, and differentiated colonies in the two different experimental conditions. Data derived from the automatic data comparison in Pluri-IQ. Results derived from two replicates. LIF, pluripotency medium; N2B27, neuronal differentiation medium.
See also Figure S2.