Abstract
The fundamental role of the brain-specific myelin transcription factor 1-like (MYT1L) gene in cases of intellectual disability and in the etiology of neurodevelopmental disorders is increasingly recognized. Yet, its function remains under-investigated. Here, we identify a network of helix–loop–helix (HLH) transcriptional regulators controlled by MYT1L, as indicated by our analyses in human neural stem cells and in the human brain. Using cell-based knockdown approaches and microarray analyses we found that (1) MYT1L is required for neuronal differentiation and identified ID1, a HLH inhibitor of premature neurogenesis, as a target. (2) Although MYT1L prevented expression of ID1, it induced expression of a large number of terminal differentiation genes. (3) Consistently, expression of MYT1L in the human brain coincided with neuronal maturation and inversely correlated with that of ID1 and ID3 throughout the lifespan. (4) Genetic polymorphisms that reduced expression of MYT1L in the hippocampus resulted in increased expression of ID1 and ID3, decreased levels of the proneural basic HLH (bHLH) transcriptional regulators TCF4 and NEUROD6 and decreased expression of genes involved in long-term potentiation and synaptic transmission, cancer and neurodegeneration. Furthermore, our neuroimaging analyses indicated that MYT1L expression associated with hippocampal volume and activation during episodic memory recall, as measured by blood-oxygen-level-dependent (BOLD) signals. Overall, our findings suggest that MYT1L influences memory-related processes by controlling a neuronal proliferation/differentiation switch of ID-bHLH factors.
Introduction
The generation of post-mitotic neurons from proliferating progenitors, their differentiation and migration to their final positions in the brain, the axonal outgrowth and the generation of functional synapses are key events required for normal brain development throughout life. Any disturbance in these co-ordinated steps can result in intellectual inability and serious developmental and neurodegenerative disorders (Vaillend et al, 2008).
The co-ordinated actions of transcriptions factors that drive tissue- and developmental stage-specific programmes of gene expression tightly regulate cell fate specification and differentiation. Progression through neurogenesis is partly determined by the sequential activities of evolutionarily conserved basic helix–loop–helix (bHLH) transcription factors. Generally, the determination genes ATOH1, NEUROD4, ASCL1, NEUROG1, and NEUROG2 initiate neurogenesis and induce the expression of the differentiation effectors NEUROD1, NEUROD2, and NEUROD6 (Sugimori et al, 2007). Given the complexity of cell fate determination processes, it is not surprising that the activity of proneural bHLH needs to be tightly regulated. To become transcriptionally active, these powerful tissue-specific transcription factors must form heterodimers with more broadly expressed bHLH proteins, or E-proteins, encoded by the TCF4, TCF3, and TCF12 genes in humans. Conversely, their function is counteracted by another class of HLH proteins, the inhibitor of differentiation (ID). These dominant negative ID proteins control cell fate determination and inhibit differentiation during development and in adult life by sequestering bHLH proteins (For a recent review, see (Wang and Baker, 2015)). ID genes have various patterns of expression, with high Id1 expression a characteristic of neural stem cells. Deletion of both Id1 and Id3 in mice results in severe phenotypes, including embryonic lethality with cardiac defects, premature withdrawal of neuroblasts from the cell cycle and premature neuronal differentiation (Lyden et al, 1999). These phenotypes of Id1−/−/Id3−/− mice, not seen in the single mutants, illustrate substantial functional overlap between these two genes.
Another transcription factor that may regulate tissue- and developmental stage-specific programmes of gene expression is myelin transcription factor 1-like (MYT1L), a brain-specific transcription factor that may be required for the expression of neuronal genes. This is suggested by the findings that the encoded protein is sufficient to convert mouse embryonic fibroblasts into functional neurons when overexpressed together with two other transcription factors, Brn2 and Ascl1 cells (Vierbuchen et al, 2010). While in this report Myt1l did not seem to be necessary for reprogramming process per se, it enhanced the differentiated phenotypes of neurons, suggesting a role for this gene in neuronal maturation. The formation of such induced neuronal cells appears to be more complicated in humans. Although overexpression of MYT1L together with BRN2 and ASCL1 could generate functional neurons from human pluripotent stem cells, further addition of NEUROD1 is necessary to convert human fibroblasts into neuronal cells (Pang et al, 2011). This highlights cell-type and species-specific effects of MYT1L and other lineage-determining transcription factors. A role for MYT1L in neural development is further supported by the identification of MYT1L mutations in patients diagnosed with various neurodevelopmental disorders. The identification of mutations on chromosome band 2p25.3 disrupting the MYT1L gene in patients with mild to moderate intellectual disability and speech delay (Bonaglia et al, 2014; de Ligt et al, 2012; De Rocker et al, 2015; Mayo et al, 2015; Stevens et al, 2011) provided the first links between haploinsufficiency of MYT1L and intellectual disability (De Rocker et al, 2015). MYT1L mutations have also been associated with diseases, such as schizophrenia (Lee et al, 2012; Vrijenhoek et al, 2008), autism (De Rubeis et al, 2014; Meyer et al, 2012), and major depressive disorder (Wang et al, 2010).
Despite such links with disease, there has been little research to date on the mechanisms by which MYT1L contributes to neuronal differentiation and its associations with relevant human brain phenotypes. To address this, we investigated the consequences MYT1L depletion on gene expression and neural differentiation in human neural stem cells in culture and in the human brain. We also measured associations between MYT1L genotypes with hippocampal structure and function using structural and functional neuroimaging. These analyses highlight a previously unrecognized MYT1L-dependant transcriptional network that offers a unique opportunity to understand the pleiotropic roles of MYT1L in human disease.
Materials and methods
Knocking-Down MYT1L in Cultured Cells
To inhibit MYT1L function in human neural stem cells, we transduced the SPC04 cell line (Desrivières et al, 2015) with lentiviruses encoding either a control short hairpin RNA (shRNA) or a shRNA against MYT1L and measured the consequences on gene expression as indicated below.
Lentiviral production and transduction of neural stem cell cultures
eGFP-containing pGIPZ lentiviral vector with the shRNAmir sequence targeting MYT1L (V2LMM_32678) or the non-silencing shRNAmir (RHS4346) were obtained from the Open Biosystem shRNAmir library (Thermofisher Scientific Open Biosystems, UK). The sense target sequences for these shRNAs were: MYT1L: 5′-CCGTGACTACTTTGACGGAAAT-3′ non-silencing control: 5′-ATCTCGCTTGGGCGAGAGTAAG-3′. Replication incompetent lentiviruses containing the MYT1L or non-silencing shRNAmir were obtained by transfecting HEK 293T cells using the Trans-lentiviral GIPZ Packaging System (Thermofisher Scientific Open Biosystems) following the manufacturer’s instructions. Supernatants containing lentiviruses were collected 48 h and 72 h post transfection, pooled, cleared by centrifugation at 1600 g at 4 °C for 10 min, filtered and ultracentrifuged at 25 000 r.p.m. for 1.5 h at 4 °C. Supernatants were carefully removed and lentiviral pellets resuspended in 30 μl of phosphate-buffered saline and aliquoted for storage at −80 °C until use. One day before transduction, SPC04 cells were seeded in 6-well laminin-coated plates at 20 000 cells/cm2. The following day, the growth media was replaced by the pre-differentiation medium in the presence of 3.5 × 105 TU/ml of lentivirus and the cells were incubated for 48 h, after which differentiation was induced. Transduction efficiency was monitored by a fluorescent microscope (Leica DMIL) with a Leica camera DFC420C (× 10 objective).
Gene expression analyses
Effects of shRNAs on MYT1L mRNA levels and on global gene expression were assessed by quantitative PCR (qPCR) and microarray analyses.
qPCR analyses
cDNAs obtained by reverse-transcription using the SuperScript III First-Strand Synthesis System (Invitrogen) following the manufacturer’s instructions, were amplified by PCR. RNA expression levels were assessed with RPL18 (60 S Ribosomal Protein L18) as internal controls for normalization, using the following forward and reverse primers: RPL18 forward 5′-GAGAGGTGTACCGGCATTTC-3′, reverse 5′-CTCTGGCACGCTCGAACT-3′ MYT1L forward 5′-TGGAGAGCAACCTGAAGACC-3′, reverse 5′-ATTCCTCTCACAGCCTGCTT-3′. PCRs were run in triplicate in the Applied Biosystems real-time PCR device (7900HT Fast Real Time PCR system) in 20 μl reactions containing 4 μl cDNA, 0.5 μM of each forward and reverse primers and 1 × Power SYBR Green Mix (Applied Biosystems) using the following cycles: 95 °C for 15 min and 40 cycles at 95 °C for 30 s and 59 °C for 30 s. The PCR reaction products were evaluated by a melting curve analysis and the PCR products quantified using the SDS software (Applied Biosystems) comparing threshold cycles (Ct). Statistical analysis was performed on the delta Ct values (ie, experimental Ct – control Ct) by ANOVA, followed by Bonferroni- or Dunnett- based post hoc analyses (α=0.05, two-sided) using the SPSS software.
Microarray analyses
Total RNA samples were processed using the TargetAmp-Nano Labeling Kit (Cambio Ltd.) and hybridized to Illumina HumanHT-12 v4 Expression BeadChips according to the manufacturers’ instructions at the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics, Oxford. Raw data extracted by the Illumina BeadStudio software were further processed in R statistical environment (http://www.r-project.org). Raw expression data were pre-processed by variance stabilizing transformation and quantile normalization using the lumi package. Non-detected probes and probes that did not vary, based on median coefficient of variation across arrays, were filtered out. Differential expression between control vs knockdown conditions was assessed using the limma package. P-values were adjusted for multiple testing according to the false discovery rate (FDR) procedure of Benjamini and Hochberg and differentially expressed genes were selected at FDR<5%. The functional annotation clustering tool, part of the Database for Annotation, Visualisation and Integrated Discovery (DAVID) web server (Huang et al, 2009) was used to determine enrichment of functional groups in set of genes mapped by differentially expressed probes (at FDR⩽10%). Clustered GO terms with PEASE⩽0.01 (ie, enrichment scores >2) were considered specifically enriched.
Gene Expression Analyses in Post-Mortem Human Brain Samples
We analyzed publicly available data sets to follow spatiotemporal gene expression patterns in the human brain and identify expression quantitative trait loci (eQTL) in MYT1L (ie, single-nucleotide polymorphisms (SNPs) associated with differences in gene expression). These are detailed in the Supplementary Methods.
Magnetic Resonance Imaging Study of the Human Hippocampus
To investigate the impact of MYT1L SNPs on hippocampus structure and function in the living human brain, we used structural magnetic resonance imaging (MRI) and an established reliable functional MRI (fMRI) neuroimaging task (Erk et al, 2010).
Human structural MRI analyses
Subjects
A sample of N=1583 adolescents (age=14.57±0.45), participants of the IMAGEN project (Schumann et al, 2010) were analyzed in this study. Written informed consent was obtained from all participants and their legal guardians.
MRI
Details of the MRI protocols and quality checks are in the Supplementary Methods. Out of 1909 images, 1583 passed these quality control checks and were used in association analyses with MYT1L genotypes.
SNP genotyping and quality controls and selection of MYT1L genotypes
DNA purification and genotyping procedures have been described previously (Desrivières et al, 2015). Ninety five SNPs contained within the MYT1L locus and its surrounding ±10 kb region were selected and the corresponding genotypes extracted from genome-wide information collected at 502 160 SNPs for 1837 subjects (900 males, 937 females) using PLINK (http://zzz.bwh.harvard.edu/plink/). This data set had been filtered to remove SNPs with call rates of <98%, minor allele frequency <1% or deviation from the Hardy–Weinberg equilibrium (P⩽1 × 10−4), and individuals with >2% missing genotypes or of unspecified sex.
Genetic association analyses
Linkage disequilibrium (LD)-based set-test association analyses were performed, testing for the additive dosage of each SNP on the volumes of the right and left hippocampus using the set-test function of PLINK, with the following parameters: LD threshold=0.5, SNP p-value threshold=0.01; maximum number of independent SNPs=5. Empirical p-value for the MYT1L set was obtained after 10 000 permutations. Gender, intracranial volume and dummy variables for scanning site were used as covariates of no interest in the analyses. Handedness and age did not influence the above associations and were not included as a covariate in our analyses. For the single SNP associations of rs17338519 with hippocampal volume, linear regression analyses were performed using the same covariates as above. Results presented for these analyses included from 1,398 individuals with non-missing status (900 males, 937 females; and 1227 right handed).
Human fMRI study of episodic memory recall
Subjects/
285 healthy adults (mean age 34.0±9.8) German volunteers with grandparents of European ancestry were recruited at Bonn, Mannheim and Berlin. Participants had no history of any neurological disorder or current psychiatric Axis I disorder including drug or alcohol dependence, as verified by SCID-I. The study was approved by the local ethics committees of the Universities of Bonn, Heidelberg and Berlin and all subjects gave written informed consent to the study. N=3 subjects were rs17338519 AA homozygotes, n=42 AC heterozygotes and n=240 CC homozygotes. The allele frequencies were in Hardy–Weinberg equilibrium. Genotype distributions did not differ between sites. Gender, age, handedness and level of education, did not differ significantly between genotype groups.
DNA-extraction and genotyping/
Genotype data for rs17338519 were extracted from a genome-wide data set generated at the Department of Genomics, Life & Brain Center, University of Bonn using Human610-Quad Bead Arrays (Illumina, San Diego, CA, USA).
Functional imaging task/
During fMRI scanning, subjects completed an episodic memory task that was part of three consecutive memory tasks, ie, encoding, recall and recognition of face-profession pairs (for a detailed description see Erk et al (2010)). During recall, the task in focus here, faces were presented together with the question whether the depicted person had to complete apprenticeship or academic studies to qualify for the respective profession that had been learned during encoding. Subjects had to indicate by button press, which qualification was correct. In the control condition, subjects had to assess whether the left or the right ear of different head contours was larger. The task consisted of 4 blocks of 4 faces (a 6 s) and 4 blocks of 6 head contours (a 4 s) each. Each block lasted 24 s. Task parameters and image processing are detailed in the Supplementary Methods.
For each subject, statistical contrast images of memory vs control were obtained and analyzed using the general linear model in a second-level random effects analysis (two-sample t-test) with site, age, gender and hippocampal volume as covariates. Given the small number of AA individuals, we pooled individuals with either one or two copies of the A allele (AA,AC). We used a region of interest (ROI) approach to investigate BOLD signals in the hippocampus, using anatomical labels provided by the Wake Forest University PickAtlas (www.fmri.wfubmc.edu/downloads).
Statistical inference/
The significance threshold was set to p<0.05, corrected for multiple comparisons within our a priori defined anatomical ROI, the hippocampus. For all analyses, we employed conservative analysis statistics by using family wise error rate (FWE), exerting strong control of type I error over multiple comparisons.
Results
Induction of MYT1L follows Neural Commitment, Correlating with the Expression of Terminal Differentiation Genes
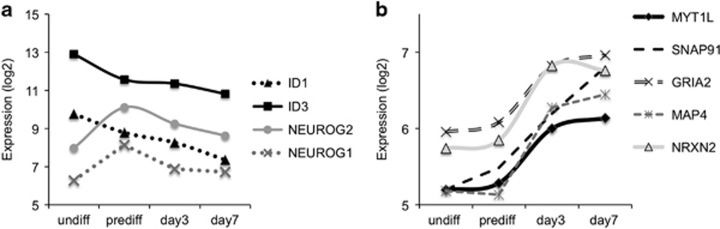
To investigate the role of MYT1L in neural differentiation, we followed its expression in SPC04, a neural progenitor cell line that efficiently differentiates under defined culture conditions (Supplementary Methods). These cells express markers of differentiated neurons and produce neurite outgrowth starting 3 days after induction of differentiation (Desrivières et al, 2015). Microarray analyses indicated that proliferating, undifferentiated SPC04 cells express high levels of the stem cells markers ID 1 and ID3. The loss of proliferation correlated with drop in expression of these genes. Conversely, expression of NEUROG1 and NEUROG2 increased when that of ID1 and ID2 decreased, being maximal at the stage preceding induction of differentiation (Figure 1a). These patterns of expression contrasted sharply with that of MYT1L, whose expression was induced with differentiation, correlating with that of markers of terminal differentiation (Figure 1b). Functional annotation clustering confirmed that expression of MYT1L positively correlated with transcripts (r>0.85; Supplementary Table 1) involved in neurite development and synaptic transmission.
Figure 1.
Changes in gene expression patterns during differentiation of the human neural stem cell line SPC04. Changes in HLH (ID1 and ID3) and bHLH (NEUROG1 and NEUROG2) mRNA levels (a), and induction of a subset of terminal differentiation genes co-expressed with MYT1L (b) were accessed by microarray analyses. Results show variations in expression in various stages of differentiation (pre-differentiation, and 3 or 7 days after induction of differentiation), relative to that of undifferentiated cells and represent mean±SEM of n=3 independent experiments.
Knockdown of MYT1L Enhances Expression of ID1 and ID3 and Inhibits Neuronal Differentiation
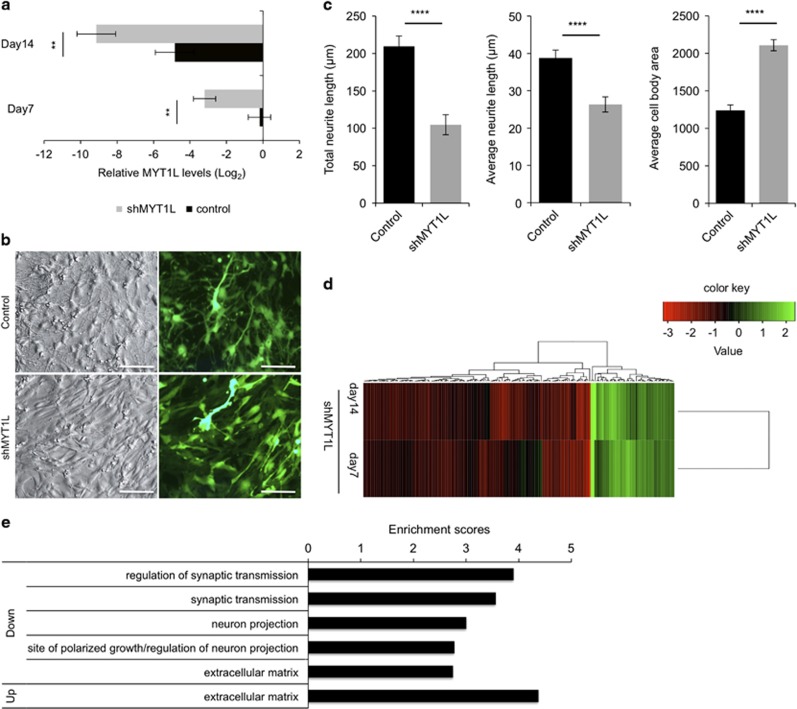
We next studied the consequences of MYT1L loss of function on neuronal differentiation and gene expression. We infected SPC04 cells with lentiviruses encoding either a shRNA against MYT1L or a control shRNA and allowed them to differentiate for 7 or 14 days. As shown in Figure 2a, treatment with shMYT1L for 7 or 14 days, resulted in significant reduction of MYT1L mRNA levels (F(1,9)=29.01, p=0.002). Morphological examination of the infected cells indicated that while infection with control lentiviruses generated cells with apparent neuronal morphology with extended networks of neurites visible 7 days after induction of differentiation, infection with shMYT1L-expressing lentiviruses prevented these morphological changes, resulting in larger cell bodies and less elaborated neurite outgrowth (Figure 2b and c; see Supplementary Methods for quantification of neurite outgrowth). We then conducted microarray analyses to identify gene networks affected by MYT1L knockdown. These analyses indicated that most transcripts that were differentially expressed upon shMYT1L treatment were downregulated (Supplementary Table 2 and Figure 2d), indicating that MYT1L acts mainly as a transcriptional activator. Functional clustering analyses indicated that genes downregulated by shMYT1L were enriched for genes involved in neurite outgrowth, axonal development, synaptic transmission and extracellular matrix composition (Figure 2e). Interestingly, the HLH ID1 and ID3 were among the few genes whose expression was induced upon MYT1L knockdown (Supplementary Table 2). These results suggested that MYT1L facilitates withdrawal from the cell cycle and commitment to neural differentiation by preventing expression of ID1 and ID3.
Figure 2.
Effects of MYT1L knockdown on neural differentiation and global gene expression in SP04 cells. Cells were transduced with lentiviral vectors encoding shMYT1L or a non-silencing shRNA control and induced to differentiate for 7 or 14 days. (a) Efficiency of MYT1L knockdown accessed by PCR 7 and 14 days after induction of differentiation. Effects of treatment with shMYT1L on MYT1L and mRNA levels, split according to the length of differentiation. Results represent mean±SEM of n=3 independent experiments; **p<0.01. (b) Effects of shMYT1L on morphology of SPC04 cells, 7 days after induction of differentiation. Changes in morphology are evident when comparing phase contrast pictures of cells transduced with control, non-silencing lentiviruses with that of cells transduced with shMYT1L-encoding lentiviruses: control cells showed enhanced neuronal features such as increased neurite elongation and retracted cell bodies. In contrast, shMYT1L prevented these morphological changes, resulting in a flat, more adherent morphology. The fluorescence pictures reveal the high efficiency of transduction by the lentiviruses that expressed the enhanced green fluorescent protein as marker. Scale bar=100 μm. (c) Quantification of the morphological changes described in (b) were obtained using the neurite outgrowth plugin in MetaMorph; total neurite length and average length of neurite per cell and cell soma area were recorded in a total of 473 cells (number of cells per group: control MYT1L, n=298; shMYT1L, n=175). Unpaired t-tests were used to compare means in each group: ****p<0.0001. (d) Microarray mRNA profiling analysis reveals target genes differentially affected by MYT1L. Probe sets differentially expressed following shMYT1L treatment were clustered together based on euclidean distance, and their expression levels (each line represents a single probe set) are displayed as fold change compared to control treatment. (e) Functional clustering of differently expressed genes showing enrichment for genes involved in neuronal maturation and the extracellular matrix composition.
Expression of MYT1L is Inversely Correlated with ID1 and ID3 Levels in the Human Brain Throughout the Lifespan
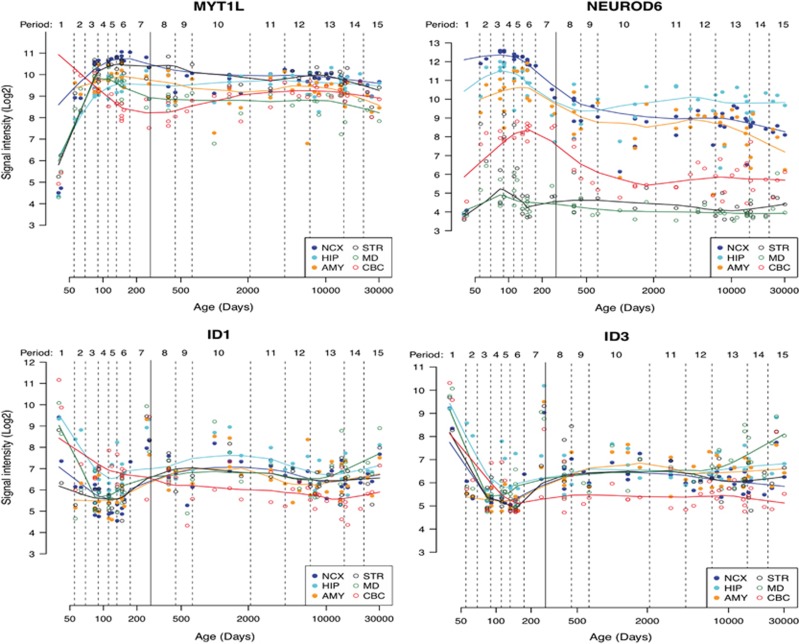
To investigate the relevance of these findings to human brain development, we analyzed publicly available data sets of human post-mortem brain samples (Supplementary Methods), comparing the expression patterns of MYT1L, ID1 and ID3 in the human brain across the lifespan (Figure 3). MYT1L is expressed throughout the human brain. Its expression increases sharply during prenatal development to reach a maximum during mid and late stages of fetal development. Thereafter, expression of MYT1L drops, remaining moderate throughout childhood and adolescence and decreasing further in adult life. Such expression profile coincides with the expression of neuronal differentiation markers throughout the lifetime (data not shown but available upon request), overlapping with that of tissue- and region-specific bHLHs, such as NEUROD6. Conversely, expression of ID1 and ID3 follows reversed patterns, being highest when MYT1L expression is lowest (ie, during embryonic development and childhood) and decreasing when MYT1L expression increased. These opposite expression patterns further support a functional antagonism between MYT1L and ID1/ID3.
Figure 3.
Analyses of gene expression in the human brain across the lifetime reveals patterns of MYT1L mRNA expression that correlates highly with that of NEUROD6, notably in the neocortex, the hippocampus and the amygdala. Conversely, MYT1L expression correlates inversely with that of ID1 and ID3 throughout the life span.
Mutations Decreasing MYT1L Expression in the Human Hippocampus Associate with Induction of ID1 and ID3 and Repression of Neural Differentiation Genes
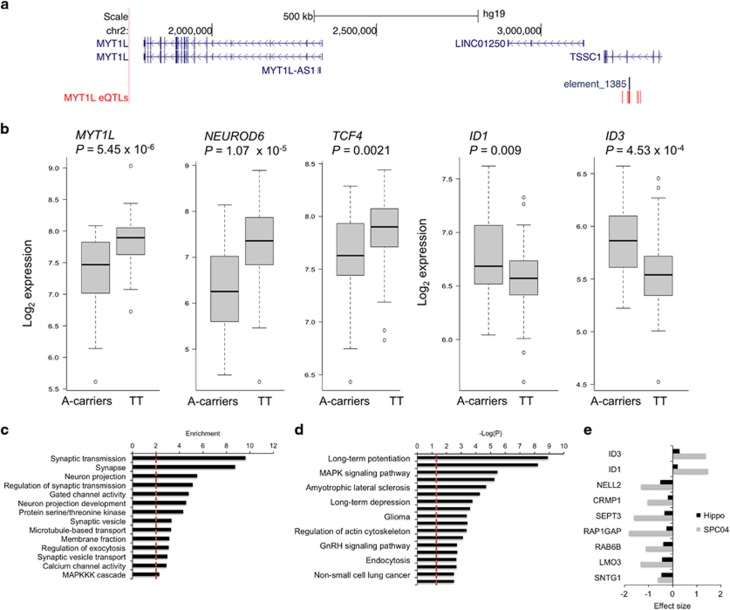
If, as suggested above, MYT1L represses expression of ID1 and ID3 in the human brain, mutations altering MYT1L functions should affect ID1 and ID3 levels such that their expression is induced when MYT1L is inhibited, or vice versa. To test this, we searched the UKBEC data set (Trabzuni et al, 2012) for potential cis-acting eQTLs, that are SNPs within the MYT1L locus that correlate with altered MYT1L mRNA levels (Supplementary Methods). We identified several such eQTLs, clustered around a conserved tissue-specific transcriptional enhancer upstream of MYT1L (Figure 4a; http://enhancer.lbl.gov/; (Visel et al, 2007)). The presence of minor alleles at these loci was associated with decreased expression of MYT1L specifically in the hippocampus. This is illustrated in Supplementary Figure 1 for rs55800610, the most significant of these eQTLs. Individuals carrying the minor A-allele at this locus had significantly lower MYT1L mRNA levels in the hippocampus (P=4.4 × 10−6). A similar, nominally significant decrease was also observed in the thalamus (P=0.042) but not in eight other brain regions. As hypothesized, the reduction of MYT1L levels in the hippocampus of carriers of the rs55800610 A-allele was accompanied with significant increased levels of ID1 and ID3 (Figure 4b).
Figure 4.
Effects of a MYT1L-associated eQTL on gene expression in the hippocampus. (a) Genomic view of the MYT1L locus, indicating SNPs acting in cis to influence MYT1L expression (MYT1L eQTLs, red bars). These SNPs are all in high linkage disequilibrium (r2>0.8), surrounding a conserved distant-acting and tissue-specific transcriptional enhancer (ie, human element hs1385, labeled as element_1385) from the VISTA Enhancer database, (Pennacchio et al, 2006). Genomic co-ordinates are based on the hg19 genome assembly. (b) The minor alleles at these eQTLs decrease mRNA levels of MYT1L (cis-effects) in the hippocampus, as illustrated for the rs55800610 gene variant. The minor A-allele at this SNP also associates with reduced NEUROD6 and TCF4 mRNA levels and increased mRNA levels of ID1 and ID3 in the hippocampus (trans-effects). Because of the low minor allele frequency (MAF=0.07), heterozygotes and individuals homozygotes for the minor allele were grouped for analyses. Genotypes count: A-carriers (AA+AT), n=20; TT, n=114. (c and d) Gene set enrichment analyses of all genes downregulated in individuals carriers of the rs55800610 A-allele indicating their clustering into specific biological functions (c) and belonging to a known biological pathway (d). The dashed vertical line represents the significance threshold (Padj 0.05). (e) Genes whose expression is affected by MYT1L decrease both in SPC04 cells and in the human hippocampus, and the measured effect sizes (negative=down-regulation, positive=up-regulation).
This SNP also affected expression of a large number of genes in trans (N=850, FDR⩽0.05), most of which (>90%), such as the proneural genes NEUROD6 and TCF4 (Figure 4b and Supplementary Table 3), were downregulated in carriers of the rs55800610 minor A-allele. The affected genes contributed largely to the regulation of synaptic transmission and long-term potentiation. Also noteworthy are significant enrichments for genes involved in intracellular signaling (calcium- and MAP kinase-related signaling pathways), in cancer (glioma, non-small cell lung cancer) and in neurodegenerative diseases like amyotrophic lateral sclerosis and Alzheimer’s disease (Figure 4c and d).
Mutation Linked to Decreased MYT1L Expression Associates with Hippocampal Volume and Hippocampal Activation During Memory Retrieval
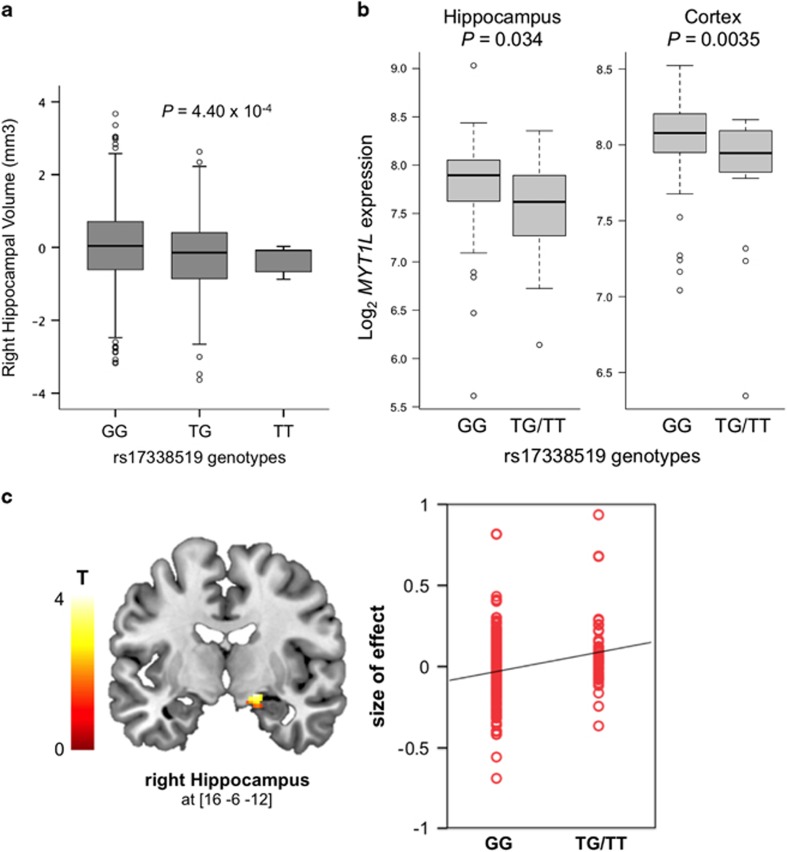
The results above suggested that MYT1L may play a role in development and function of the hippocampus. We investigated this further by analyzing associations between MYT1L SNPs and structural and fMRI in the human hippocampus. We first analyzed possible associations between MYT1L and inter-individual variations of hippocampal volume in IMAGEN, a cohort of N=1,398 healthy adolescents, whose characteristics are listed in Supplementary Table 4. As rs55800610 was not directly genotyped in IMAGEN, we performed gene set-based analyses with SNPs genotyped in this sample to identify independent SNPs associating with hippocampal volume. For this, 95 SNPs contained within the MYT1L locus and its surrounding±10 kb region were tested for association by linear regression analyses taking LD between SNPs into account. Five SNPs in high LD were associated with right hippocampal volume below a p-value of 0.01. Only 1 of these, rs17338519, was independently significant based on an r2 threshold of 0.5, remaining significant after 10 000 permutations (empirical p-value=0.0098). For the left hippocampus, two SNPs in high LD (r2>0.5), including rs17338519, were associated with hippocampal volume below a p-value threshold of 0.01. However, this association did not remain significant after 10 000 permutations (empirical p-value=0.3598). Linear regression analyses revealed significant association between genotypes at rs17338519 and right hippocampal volume. As shown in Figure 5a, the volume of the right hippocampus significantly decreased with the number of minor T-alleles at this locus (β=−75.96, p=4.40 × 10−4, Figure 5a). Although no SNP passed the threshold of significance for analyses on the left hippocampus, we noted that the effects of rs17338519 on the right and left hippocampus were in the same direction (β=−55.55, p=0.0079, for effects of rs17338519 on the left hippocampus). We also used the UKBEC data set to test whether genotypes at rs17338519 were related to MYT1L expression levels in the human brain. Regression analyses indicated that the presence of the T-allele associated with decreased MYT1L mRNA levels in the hippocampus and the cortex (β=−0.22, p=0.034; β=−0.22, p=0.0035; Figure 5b).
Figure 5.
Associations between MYT1L expression and hippocampus volume and hippocampus activation during episodic memory recall. (a) Association of rs17338519 with hippocampal volume in a sample of N=1,398 adolescents. Linear regression analyses indicated that the number of minor T-alleles associated with lower volume of the right hippocampus (P=4.40 × 10−4). (b) Expression of MYT1L stratified by genotypes at rs17338519, in the hippocampus and cerebral cortex of 134 post-mortem human brains. For the cortex, expressions of MYT1L in the occipital, temporal, and frontal cortices were averaged. Regression analyses indicated that the presence of the T-allele at rs17338519 associated with decreased MYT1L mRNA levels in the hippocampus (P=0.034) and the cortex (P=0.0035). Genotypes count: GG=110; T-carriers=24. (c) Association of rs17338519 with hippocampal activation during an fMRI task of episodic memory recall in 285 healthy adults. The left panel shows the ROI-derived brain activation map within the hippocampus, with increased activation of the right hippocampus (x=16, y=−6, z=−12) during episodic memory recall in carriers of the T-allele (N=285; p=0.02 FWE corrected for multiple testing across ROI). The right panel shows a quantification of rs17338519 genotypes effects on the activation of the right hippocampus. Each dot represents size of effect in one subject.
To investigate a possible link between MYT1L and hippocampal function, we used an established reliable neuroimaging task (Erk et al, 2010), testing for the effects of rs17338519 on hippocampus activation during episodic memory recall in N=285 healthy individuals. We observed increased activation of the right hippocampus (x=16, y=−6, z=−12; Z=3.65, p=0.022 FWE corrected for ROI) during memory recall in carriers of the T-allele (Figure 5c). Genotype groups in this sample did not differ significantly for memory encoding or recognition. These data suggest that MYT1L maintains hippocampal structural integrity and its activation during learning and memory.
Discussion
We have shown for the first time that the MYT1L transcription factor regulates neurogenesis by repressing ID genes ID1 and ID3, and inducing proneural bHLH genes such as TCF4, NEUROD2, and NEUROD6. Loss-of-function experiments in vitro and the identification of deleterious mutations in the human brain indicated that this transcriptional regulator is necessary for the expression of genes implicated several aspects of neural development such as neurite outgrowth, axonal development and the regulation of synaptic transmission, as well as extracellular matrix composition. In line with a role for ID proteins in cell cycle and senescence (Ruzinova and Benezra, 2003), MYT1L-regulated genes were also enriched for genes involved in cancer and the neurodegenerative diseases amyotrophic lateral sclerosis and Alzheimer’s disease. Neuroimaging analyses indicated that expression of MYT1L in the hippocampus associates with hippocampus volume and activation during episodic memory recall. Thus, we propose that MYT1L controls neural development and influences cognitive capacity by displacing the balance of bHLH and ID factors such that bHLH predominate.
The links that we identified between MYT1L and ID gene expression are interesting. Id proteins control proliferation and differentiation of a variety of tissue-specific stem cells in response to signaling pathways triggered by their extracellular microenvironment (Chaudhary et al, 2001; Jankiewicz et al, 2006; Niola et al, 2012; Ying et al, 2003; Yokota et al, 1999). In this context, one notable finding is the link that we established between MYT1L and extracellular matrix gene expression. Interestingly, RAP1GAP, a gene downregulated by loss of function of MYT1L, both in SPC04 cells and in the hippocampus (Figure 4e and Supplementary Tables 2 and 3), is known as a direct target of the bHLH—Id transcriptional pathway that encodes a key regulator of cell adhesion, as determined by investigations in the developing mouse brain. Id-mediated repression of Rap1GAP has been found to enhance adhesion of neural stem cells to their extracellular microenvironment, thereby synchronizing neural stem cell functions to residency in the specialized stem cell niche (Niola et al, 2012). Therefore, MYT1L-mediated negative control of ID expression may activate a transcriptional program to synchronize the committed differentiation of neural stem cell progenitors and their detachment from the stem cell niche. Consistent with this role in regulating stem cell anchorage to the niche and neural differentiation, ID1/RAP1GAP has a central role in tumor formation, particularly in the formation of highly aggressive glioblastomas (Niola et al, 2013; Soroceanu et al, 2013). Our results also implicate MYT1L in glioma formation, through a process that besides RAP1GAP, may require MAP kinases and calcium-dependent signaling mechanisms (Ren et al, 2007). Indeed, the MYT1L gene was found to be deleted in ~5% of glioblastoma multiforme samples and the protein absent or downregulated in >80% of these samples (Hu et al, 2013), leading to the suggestion that gliomagenesis requires neutralization of terminal differentiation.
Importance of the MYT1L-ID-HLH network for normal brain development and function is reflected by the severe developmental delays and complex phenotypes associated with disruptions in some of its core components. Although MYT1L mutations have been associated with intellectual disability with early onset obesity (Doco-Fenzy et al, 2014; Stevens et al, 2011), autism (Meyer et al, 2012; Rio et al, 2013), and schizophrenia (Lee et al, 2012; Vrijenhoek et al, 2008), TCF4 mutations cause Fuchs’corneal endothelial dystrophy, Pitt–Hopkins syndrome, a disorder that is part of the clinical spectrum of Rett-like syndromes characterized by intellectual disability and developmental delay (Whalen et al, 2012), and have been described in patients with schizophrenia, autism, and other neurodevelopmental disorders (Forrest et al, 2014). Strinkingly, expression of ID genes is also linked to Rett syndrome (Wang and Baker, 2015). These genes are repressed by MeCP2, a gene required for maturation of neurons and whose defect causes Rett syndrome, mental retardation, encephalopathy, and autism susceptibility (Gao et al, 2015; Peddada et al, 2006). Such results indicate that differential regulation of the ID-HLH pathway by MYT1L may underlie the phenotypic overlap of several neurodevelopmental disorders.
The identification of genetic variants affecting MYT1L expression in the hippocampus had guided our MRI studies on the hippocampus. The location of the rs55800610 variant within a highly conserved tissue-specific enhancer that directs spatially restricted gene expression in the developing brain might underlie the restricted effect of this SNP in the hippocampus, as rs17338519 that is not located in such regulatory region has broader effects in the brain. Finding such functions for MYT1L in the hippocampus is in agreement with this region being one of the few in the mammalian brain that retains its ability to produce functional neurons throughout life. Besides its major role in postnatal neurogenesis (Schwarz et al, 2012) the hippocampus is crucial for episodic memory (Bliss and Collingridge, 1993) and changes in hippocampal volumes are hallmarks of several common neurological and psychiatric disorders including mental retardation (Nadel, 2003). Yet, the hippocampus is not the only key brain area for memory and more generally, cognitive ability. Structural and functional neuroimaging have revealed a network of cortical brain areas involved in general cognitive functions (Deary et al, 2010). Our findings that the minor allele at rs17338519, that affects hippocampal activation during memory recall, is linked with decreased MYT1L expression in both hippocampus and cortex are in agreement with this.
It is remarkable that the learning and memory deficits of subjects with intellectual disability are associated with decreased size of the hippocampus and impairments of the prefrontal cortex and hippocampal system. These deficits become considerably more noticeable during late childhood and adolescence (Nadel, 2003), possibly as genetic factors underlying inter-individual variability in these brain systems become effective at this stage of development (Haworth et al, 2010). Our data suggest that MYT1L is one of these factors underlying inter-individuals differences in hippocampal volume. The increased hippocampal activation observed in our fMRI study also suggests that decreased MYT1L levels may lessen neuronal efficiency during cognitive task performance. However, we acknowledge that the use of two distinct cohorts of different ages for our MRI analyses has limitations. Future longitudinal studies of cohorts with structural MRI and suitable fMRI assessments are required to determine if genetic alterations in MYT1L predispose to changes in brain structure and function across the lifetime. Nonetheless, the discoveries described here provide etiologically relevant mechanistic explanations for a role of MYT1L in neural development.
Data Access
The microarray data generated for this publication have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE65945 and GSE65857.
Funding and disclosure
This work was funded by Wellcome Trust grant reference 090532/Z/09/Z and MRC Hub grant G0900747 91070. This work received further support from the European Union-funded FP6 Integrated Project IMAGEN (reinforcement-related behavior in normal brain function and psychopathology; LSHM-CT- 2007-037286). S.D and G.S. are also supported in part by the NIH BD2K award, U54EB020403, the FP7 projects IMAGEMEND (602450; IMAging GEnetics for MENtal Disorders) and MATRICS (603016), the Innovative Medicine Initiative Project EU-AIMS (115300-2), a Medical Research Council Programme Grant ‘Developmental pathways into adolescent substance abuse’ (93558), the Swedish funding agency FORMAS, the Medical Research Council and the Wellcome Trust (Behavioural and Clinical Neuroscience Institute, University of Cambridge), the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, the Bundesministeriumfür Bildung und Forschung (BMBF grants 01GS08152; 01EV0711; eMED SysAlc01ZX1311A; Forschungsnetze AERIAL and BipoLife) and the Deutsche Forschungsgemeinschaft (DFG grants FOR 1617, SFB 940 and SM 80/5-2). The authors declare no conflict of interest.
Acknowledgments
We thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics for the generation of the Gene Expression data. A.K. and L.M.M were recipients of a studentship form the Medical Research Council, UK and Consejo Nacional de Ciencia y Tecnología (CONACyT; México), respectively.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Bliss TV, Collingridge GL (1993). A synaptic model of memory: long-term potentiation in the hippocampus. Nature 361: 31–39. [DOI] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Zanini S (2014). A new patient with a terminal de novo 2p25.3 deletion of 1.9 Mb associated with early-onset of obesity, intellectual disabilities and hyperkinetic disorder. Mol Cytogenet 7: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary J, Johnson J, Kim G, Skinner MK (2001). Hormonal regulation and differential actions of the helix-loop-helix transcriptional inhibitors of differentiation (Id1, Id2, Id3, and Id4) in Sertoli cells. Endocrinology 142: 1727–1736. [DOI] [PubMed] [Google Scholar]
- de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T et al (2012). Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 367: 1921–1929. [DOI] [PubMed] [Google Scholar]
- De Rocker N, Vergult S, Koolen D, Jacobs E, Hoischen A, Zeesman S et al (2015). Refinement of the critical 2p25.3 deletion region: the role of MYT1L in intellectual disability and obesity. Genet Med 17: 460–466. [DOI] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE et al (2014). Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515: 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deary IJ, Penke L, Johnson W (2010). The neuroscience of human intelligence differences. Nat Rev Neurosci 11: 201–211. [DOI] [PubMed] [Google Scholar]
- Desrivières S, Lourdusamy A, Tao C, Toro R, Jia T, Loth E et al (2015). Single nucleotide polymorphism in the neuroplastin locus associates with cortical thickness and intellectual ability in adolescents. Mol Psychiatry 20: 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doco-Fenzy M, Leroy C, Schneider A, Petit F, Delrue MA, Andrieux J et al (2014). Early-onset obesity and paternal 2pter deletion encompassing the ACP1, TMEM18, and MYT1L genes. Eur J Hum Genet 22: 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erk S, Meyer-Lindenberg A, Schnell K, Opitz von Boberfeld C, Esslinger C, Kirsch P et al (2010). Brain function in carriers of a genome-wide supported bipolar disorder variant. Arch Gen Psychiatry 67: 803–811. [DOI] [PubMed] [Google Scholar]
- Forrest MP, Hill MJ, Quantock AJ, Martin-Rendon E, Blake DJ (2014). The emerging roles of TCF4 in disease and development. Trends Mol Med 20: 322–331. [DOI] [PubMed] [Google Scholar]
- Gao H, Bu Y, Wu Q, Wang X, Chang N, Lei L et al (2015). Mecp2 regulates neural cell differentiation by suppressing the Id1 to Her2 axis in zebrafish. J Cell Sci 128: 2340–2350. [DOI] [PubMed] [Google Scholar]
- Haworth CM, Wright MJ, Luciano M, Martin NG, de Geus EJ, van Beijsterveldt CE et al (2010). The heritability of general cognitive ability increases linearly from childhood to young adulthood. Mol Psychiatry 15: 1112–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Ho AL, Yuan L, Hu B, Hua S, Hwang SS et al (2013). From the Cover: neutralization of terminal differentiation in gliomagenesis. Proc Natl Acad Sci USA 110: 14520–14527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Jankiewicz M, Groner B, Desrivières S (2006). Mammalian target of rapamycin regulates the growth of mammary epithelial cells through the inhibitor of deoxyribonucleic acid binding Id1 and their functional differentiation through Id2. Mol Endocrinol 20: 2369–2381. [DOI] [PubMed] [Google Scholar]
- Lee Y, Mattai A, Long R, Rapoport JL, Gogtay N, Addington AM (2012). Microduplications disrupting the MYT1L gene (2p25.3) are associated with schizophrenia. Psychiatr Genet 22: 206–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O'Reilly R et al (1999). Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature 401: 670–677. [DOI] [PubMed] [Google Scholar]
- Mayo S, Rosello M, Monfort S, Oltra S, Orellana C, Martinez F (2015). Haploinsufficiency of the MYT1L gene causes intellectual disability frequently associated with behavioral disorder. Genet Med 17: 683–684. [DOI] [PubMed] [Google Scholar]
- Meyer KJ, Axelsen MS, Sheffield VC, Patil SR, Wassink TH (2012). Germline mosaic transmission of a novel duplication of PXDN and MYT1L to two male half-siblings with autism. Psychiatr Genet 22: 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadel L (2003). Down's syndrome: a genetic disorder in biobehavioral perspective. Genes Brain Behav 2: 156–166. [DOI] [PubMed] [Google Scholar]
- Niola F, Zhao X, Singh D, Castano A, Sullivan R, Lauria M et al (2012). Id proteins synchronize stemness and anchorage to the niche of neural stem cells. Nat Cell Biol 14: 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niola F, Zhao X, Singh D, Sullivan R, Castano A, Verrico A et al (2013). Mesenchymal high-grade glioma is maintained by the ID-RAP1 axis. J Clin Invest 123: 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ et al (2011). Induction of human neuronal cells by defined transcription factors. Nature 476: 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peddada S, Yasui DH, LaSalle JM (2006). Inhibitors of differentiation (ID1, ID2, ID3 and ID4) genes are neuronal targets of MeCP2 that are elevated in Rett syndrome. Hum Mol Genet 15: 2003–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M et al (2006). In vivo enhancer analysis of human conserved non-coding sequences. Nature 444: 499–502. [DOI] [PubMed] [Google Scholar]
- Ren H, Yang BF, Rainov NG (2007). Receptor tyrosine kinases as therapeutic targets in malignant glioma. Rev Recent Clin Trials 2: 87–101. [DOI] [PubMed] [Google Scholar]
- Rio M, Royer G, Gobin S, de Blois MC, Ozilou C, Bernheim A et al (2013). Monozygotic twins discordant for submicroscopic chromosomal anomalies in 2p25.3 region detected by array CGH. Clin Genet 84: 31–36. [DOI] [PubMed] [Google Scholar]
- Ruzinova MB, Benezra R (2003). Id proteins in development, cell cycle and cancer. Trends Cell Biol 13: 410–418. [DOI] [PubMed] [Google Scholar]
- Schumann G, Loth E, Banaschewski T, Barbot A, Barker G, Buchel C et al (2010). The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Mol Psychiatry 15: 1128–1139. [DOI] [PubMed] [Google Scholar]
- Schwarz TJ, Ebert B, Lie DC (2012). Stem cell maintenance in the adult mammalian hippocampus: a matter of signal integration? Dev Neurobiol 72: 1006–1015. [DOI] [PubMed] [Google Scholar]
- Soroceanu L, Murase R, Limbad C, Singer E, Allison J, Adrados I et al (2013). Id-1 is a key transcriptional regulator of glioblastoma aggressiveness and a novel therapeutic target. Cancer Res 73: 1559–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens SJ, van Ravenswaaij-Arts CM, Janssen JW, Klein Wassink-Ruiter JS, van Essen AJ, Dijkhuizen T et al (2011). MYT1L is a candidate gene for intellectual disability in patients with 2p25.3 (2pter) deletions. Am J Med Genet A 155A: 2739–2745. [DOI] [PubMed] [Google Scholar]
- Sugimori M, Nagao M, Bertrand N, Parras CM, Guillemot F, Nakafuku M (2007). Combinatorial actions of patterning and HLH transcription factors in the spatiotemporal control of neurogenesis and gliogenesis in the developing spinal cord. Development 134: 1617–1629. [DOI] [PubMed] [Google Scholar]
- Trabzuni D, Wray S, Vandrovcova J, Ramasamy A, Walker R, Smith C et al (2012). MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Hum Mol Genet 21: 4094–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillend C, Poirier R, Laroche S (2008). Genes, plasticity and mental retardation. Behav Brain Res 192: 88–105. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463: 1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA (2007). VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res 35(Database issue): D88–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E, Sabatti C, Geurts van Kessel A et al (2008). Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet 83: 504–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Baker NE (2015). E proteins and ID proteins: helix-loop-helix partners in development and disease. Dev Cell 35: 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zeng Z, Li T, Liu J, Li J, Li Y et al (2010). Common SNPs in myelin transcription factor 1-like (MYT1L): association with major depressive disorder in the Chinese Han population. PLoS ONE 5: e13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen S, Heron D, Gaillon T, Moldovan O, Rossi M, Devillard F et al (2012). Novel comprehensive diagnostic strategy in Pitt-Hopkins syndrome: clinical score and further delineation of the TCF4 mutational spectrum. Hum Mutat 33: 64–72. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A (2003). BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115: 281–292. [DOI] [PubMed] [Google Scholar]
- Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S et al (1999). Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature 397: 702–706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.