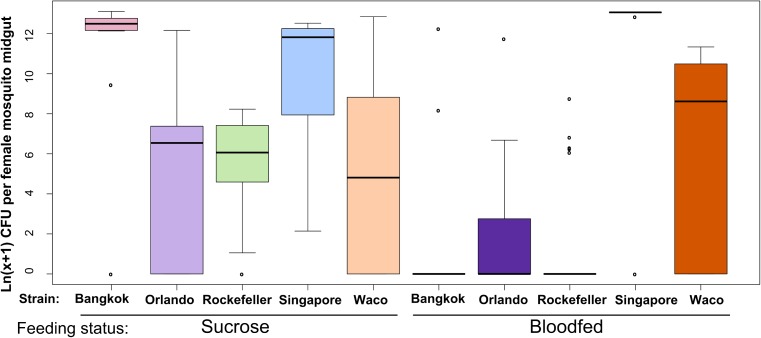
Fig 1. A. aegypti strains vary significantly in their LB-cultivable gut microbial load.
We reared female A. aegypti from five strains in parallel and mixed the breeding water between strains three times during larval/pupal development. We maintained adults on 3% sucrose until 5 days post eclosion, at which point a subset of the females were given a sterile blood meal. At 48 hours post blood feeding, we externally sterilized all adults with 70% EtOH, dissected midguts from each strain/feeding treatment, homogenized individual midguts in 1X PBS using a sterile pestle and diluted the homogenate 1:100 using additional 1X PBS. We spread 80ul of undiluted and 1:100 diluted homogenate on LB agar plates and allowed the plates to grow for 48 hours at room temperature. We combined counts from all bacterial genera found in each individual to determine total CFUs per individual female midgut. We collected data over two experimental replicates; sample sizes are as follows: Bkksucrose: n = 13, Bkkblood: n = 11, Orlsucrose: n = 18, Orlblood: n = 24, Rocksucrose: n = 18, Rockblood: n = 25, Singsucrose: n = 13, Singblood: n = 11, Wacosucrose: n = 7, Wacoblood: n = 8. For many individuals, zero bacterial colonies grew on the LB plates, which resulted in this dataset being zero-inflated. We therefore performed a zero-inflated data analysis to test for the effect of strain, feeding status and a strain × feeding status interaction on the presence/absence of bacteria or total bacterial load. We then performed stepwise backwards selection of model terms using likelihood ratio tests to assess the significance of each dropped term. For count data (i.e. the part of the model analyzing individuals with at least one CFU), strain was a significant predictor of bacterial load (p = 6.19 x 10−14) and we detected no interaction between strain and feeding status. For presence/absence data (i.e. the part of the model analyzing the presence or absence of bacteria), we detected a significant interaction (p = 0.01739) between strain and feeding status.