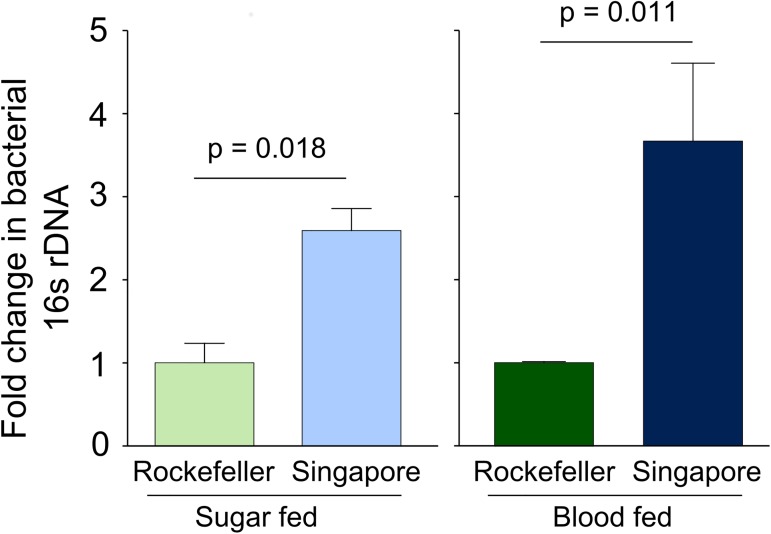
Fig 3. Rockefeller midguts have relatively lower bacterial 16S gene levels compared to Singapore midguts as measured by qPCR.
We reared female Rockefeller and Singapore strain A. aegypti in parallel and mixed the breeding water between the strains three times during larval/pupal development. Adults were maintained on 3% sucrose upon eclosion. At 3–5 days post eclosion, females were either provided a sterile blood meal or sterile sucrose. Twenty-four hours post blood meal, we externally sterilized all females by washing with 70% EtOH and then dissected two pools of 8 midguts in sterile 1X PBS for each strain and feeding treatment (5–6 midguts were collected for 3 Singapore blood fed samples). Sugar fed individuals were collected from three independent replicate experiments and blood fed individuals were collected from two. We extracted DNA from each midgut pool and used qPCR to quantify levels of the bacterial16S rDNA gene and A. aegypti S7 reference gene. We averaged delta Ct values from pools from the same biological replicate before analysis to prevent pseudoreplication. Values in the figure were calculated using the delta delta Ct method, where Singapore is shown relative to Rockefeller within each feeding treatment. Error bars represent one standard error. Raw delta Ct values were analyzed in R by ANOVA followed by a Tukey’s test using the following model: Yijk = μ + strainj + feeding statusk + strainj * feeding statusk. Both strain (p = 0.00064) and feeding status (p = 3.22 x 10−6) were highly significant and we failed to detect an interaction between strain and feeding status.