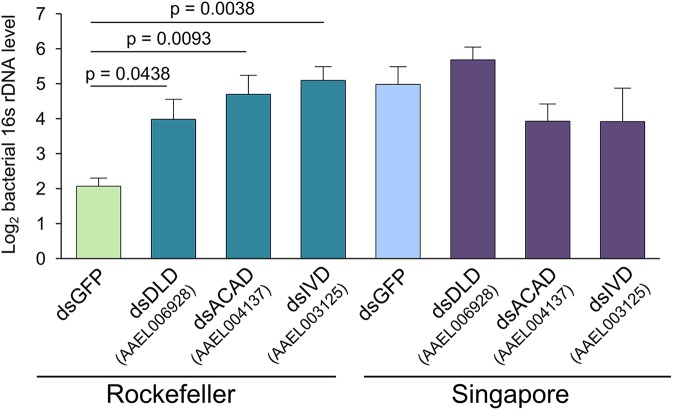
Fig 6. RNAi silencing of genes involved in Val-Leu-Ile degradation causes Rockefeller-specific increases in relative bacterial 16S rDNA.
We reared Rockefeller and Singapore mosquitoes in parallel and injected them at 3–5 days post-eclosion with 200ng of dsRNA targeting one of the three experimental genes or eGFP as a control. At two days post injection, we dissected two pools of eight midguts for each strain/treatment combination. This entire experiment was repeated three independent times. We extracted DNA from each midgut pool and performed qPCR to quantify levels of the bacterial16S rDNA gene and A. aegypti S7 reference gene. We averaged delta Ct values from pools from the same biological replicate before analysis to prevent pseudoreplication, and Y-axis values are average inverse delta CT values, i.e. -1*(CT16S –CTS7) for each treatment. Because CT values are Log2, a difference of 1 on the y-axis corresponds to a 2-fold change in 16S DNA abundance. Error bars represent one standard error. Raw delta CT values were analyzed in R by ANOVA followed by a Dunnett’s test using the following model: Yijk = μ + strainj + treatmentk + strainj * treatmentk. We detected a significant interaction between strain and treatment (p = 0.0044), and using a Dunnett’s test found that silencing all three genes caused a significant increase in 16S rRNA gene levels relative to Rock GFP-injected controls. No significant effects of silencing were detected in Sing females. DLD = dihydrolipoamide dehydrogenase, ACAD = acyl-CoA dehydrogenase, IVD = isovaleryl-CoA dehydrogenase.