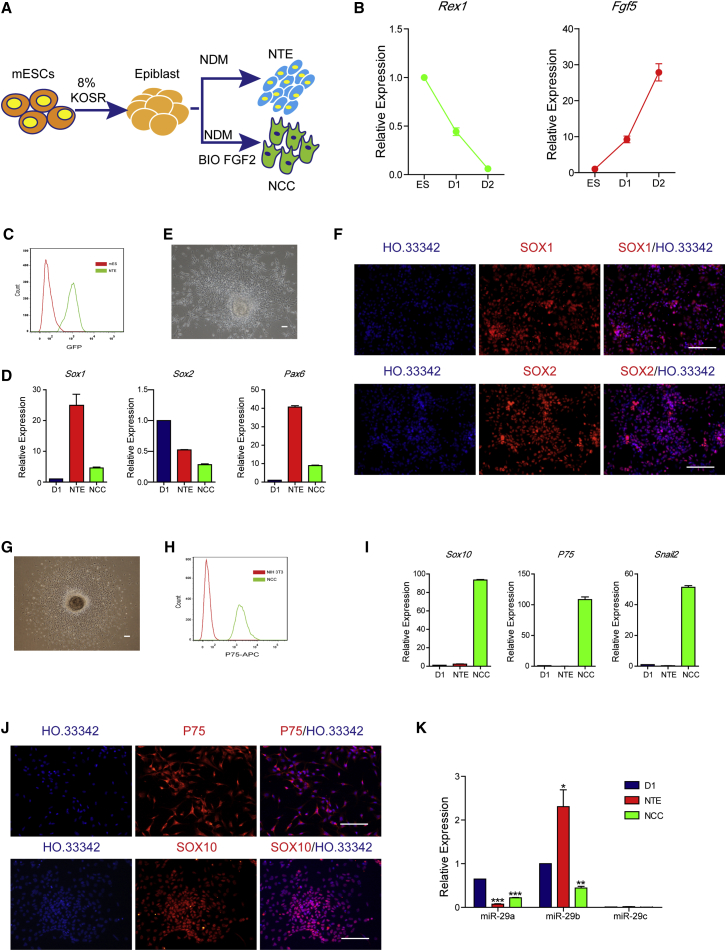
Figure 1.
MiR-29b Exhibits a Discriminating Expression Level between NTE Cells and NCCs
(A) Schematic showing the procedure for mESC differentiation into NTE and NCC.
(B) The expression level of Rex1 was downregulated and that of Fgf5 was upregulated as verified by qPCR during the differentiation from embryonic stem cell (ES) to D2.
(C) FACS analyzed the positive ratio of SOX1-GFP of mESC-NTE cells (green line) and undifferentiated ESCs (red line).
(D) The neural lineage-associated genes Sox1, Sox2, and Pax6 were upregulated as verified by qPCR in NTE cells.
(E) The epithelial cells were observed after NTE EBs had attached to a Matrigel-coated surface.
(F) Immunofluorescence assays of SOX1 and SOX2 in NTE cells.
(G) The mesenchymal-like cells were observed to migrate out of the spheres after NCC EBs had attached to a Matrigel-coated surface.
(H) FACS analyzed the positive ratio of P75 of mESC-NCCs (green line) and NIH-3T3 (red line).
(I) The neural crest-associated genes Sox10, P75, and Snail2 were upregulated as verified by qPCR in NCCs.
(J) Immunofluorescence assays of P75 and SOX10 in NCCs.
(K) qPCR measured the expression levels of miR-29 family of the NTE cells, NCCs, and D1 EBs.
Means ± SEM from n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus the control. Scale bars, 100 μm.