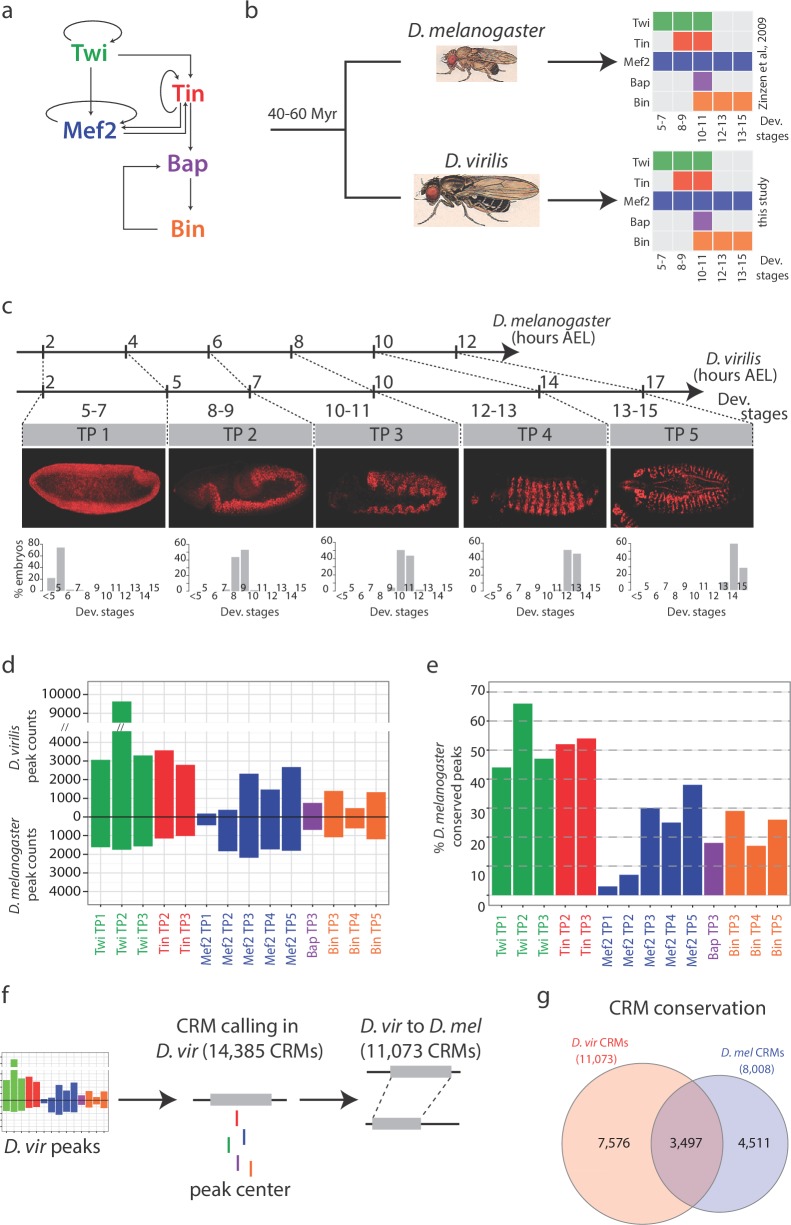
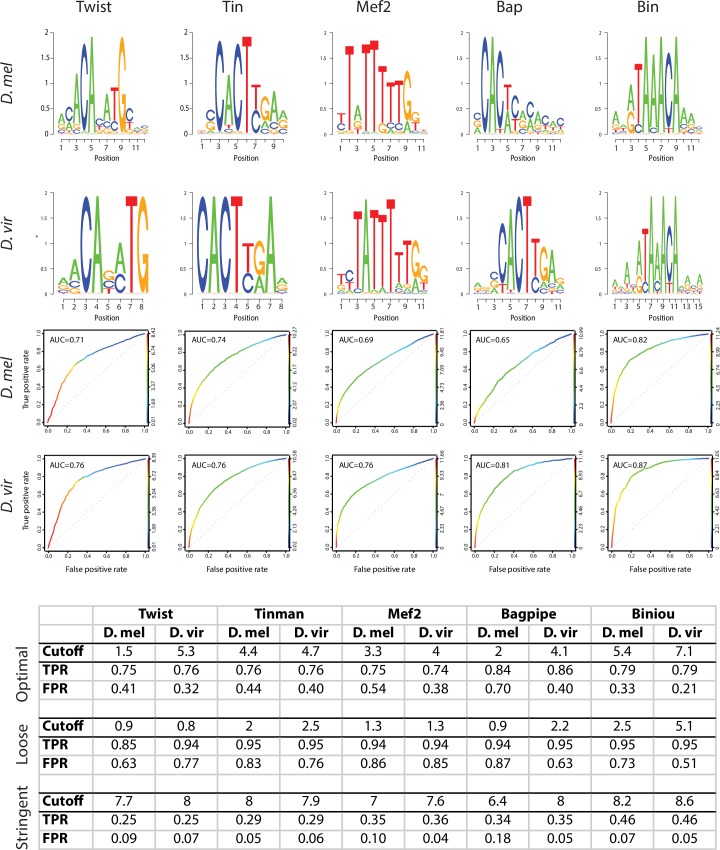
Figure 1. Conservation in TF occupancy across developmental and evolutionary time-scales.
(a) The regulatory network linking 5 mesodermal transcription factors essential for mesoderm specification. (b) D. melanogaster and D. virilis ChIP were performed using species-specific antibodies for Twi, Tin, Bap and Bin. Colored boxes illustrate TFs and time points analyzed, with developmental stages on the x-axis. (c) The scale illustrates equivalent time points for corresponding stages between D. melanogaster and D. virilis. Axis labels correspond to hours of development, after egg laying (AEL) and numbers below axis correspond to developmental stages and Time Points (TP 1–5). Confocal images of embryos with Mef2 immuno-stains at the most representative stage for each time-point, orientated anterior-left, dorsal-up. Barplots show percentage of embryos at each stage (x-axis) for each TP, averaged over two replicates. (d) Number of significant ChIP peaks in each condition in both species. TP1, TP2, etc, correspond to Time point 1, Time point 2 etc. D. melanogaster peaks are from (Zinzen et al., 2009). (e) Percentage of conserved peaks between D. melanogaster and D. virilis. (f) Peak summits in close proximity were used to define cis-regulatory modules (CRMs, see Materials and methods). (g) Venn diagram of orthologous CRMs conserved between both species; 3,497 CRMs (D. mel CRMs overlapping more than one D. vir CRMs are counted once).