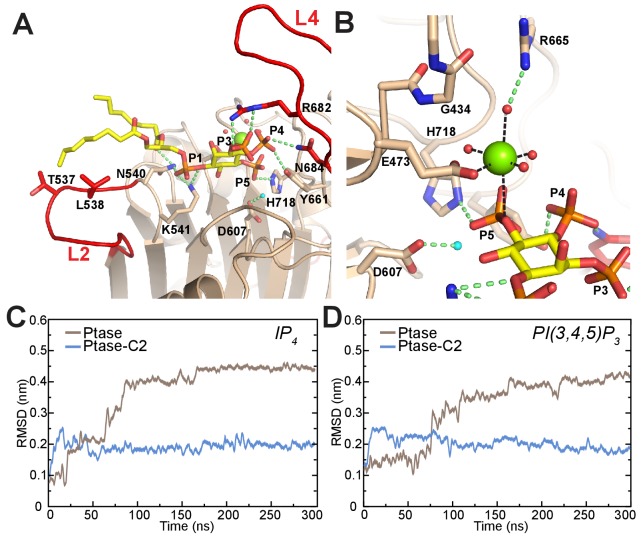
Figure 6. Model of substrate bound SHIP2.
(A) The SHIP2 Ptase-PI(3,4,5)P3-diC8 complex is modeled based on crystal structures of the homologue INPP5B crystal structure bound to PI(4)P (pdbs: 3MTC). L4 is in the ‘in’ conformation and R682 makes hydrogen bonds to the PI(3,4,5)P3 3-phosphate (P3). N540 and K541 interact with P1, N684 and Y661 with P4, H718 with P5 and L2 with PI(3,4,5)P3 lipid chains. The attacking water, bound to D607, is colored light blue and the Mg2+ ion is shown as green sphere. (B) Close-up of the Mg2+ coordination. (C–D) Plotted are RMSDs of the substrate headgroup atoms compared to the starting positions during MD simulations of the SHIP2 Ptase or Ptase-C2 bound to IP4 (C) or PI(3,4,5)P3-diC8 (D). The simulations are started with substrate positions according to the model shown in panel (A).