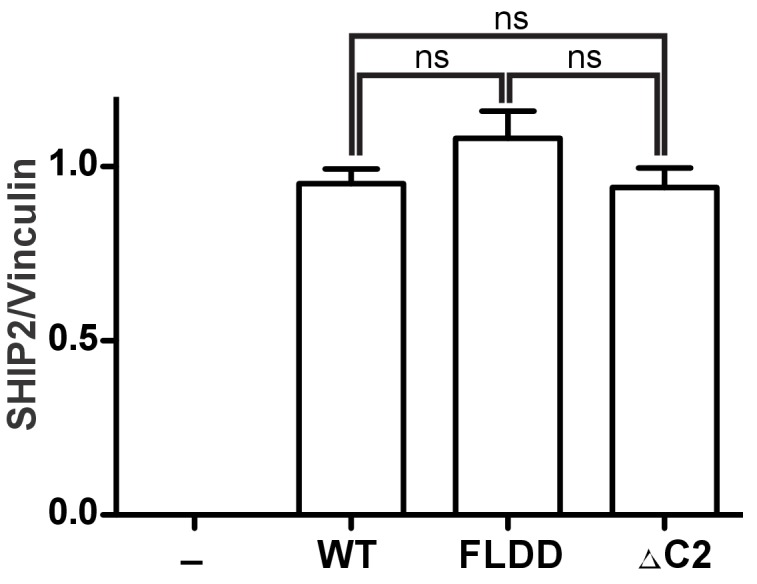
Figure 8. SHIP2 cellular activity.
Full-length SHIP2 proteins were transiently expressed in HEK293 cells and resulting Akt-pT308 levels monitored. Shown are transfections of empty vector (-), wild type SHIP2 (WT), the FLDD mutant and C2 deleted SHIP2 (ΔC2). A typical blot is shown and quantifications of blots are averaged from eight independent experiments, each in triplicates (n = 24), with SEM’s indicated. *p<0.05; ****p<0.0001 (unpaired Student t test).
DOI: http://dx.doi.org/10.7554/eLife.26640.029
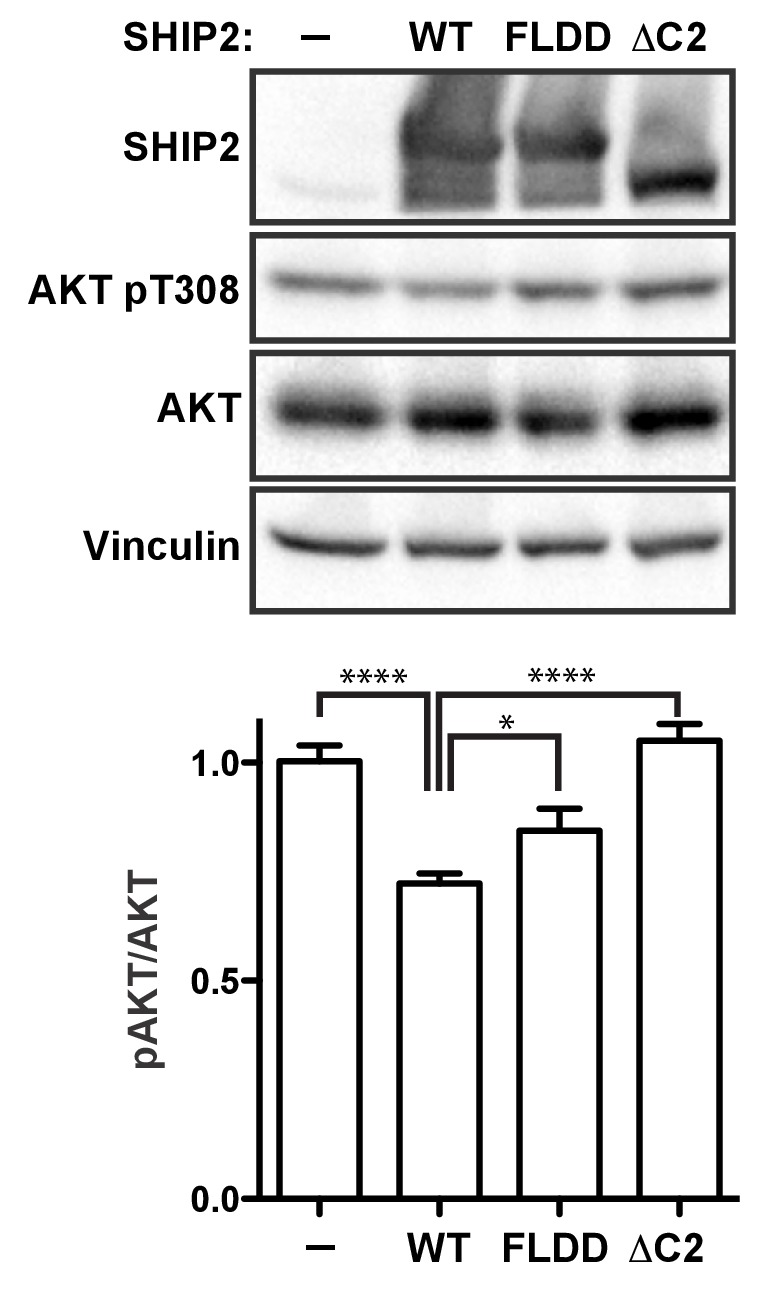
Figure 8—figure supplement 1. Quantification of SHIP expression levels.