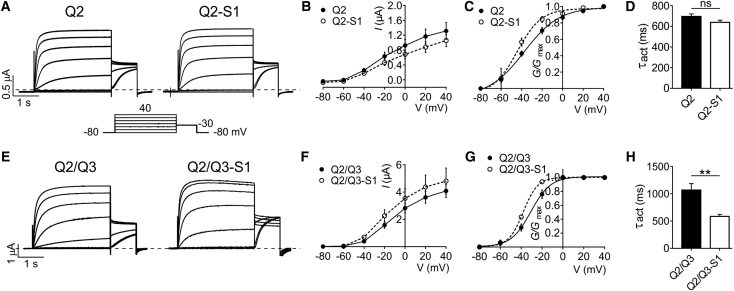
Figure 2.
SMIT1 negative-shifts and speeds KCNQ2/3 activation. (A) Representative traces from TEVC recordings of Xenopus laevis oocytes injected with cRNA encoding KCNQ2, alone or co-injected with cRNA encoding SMIT1; n = 29–32. (B) Mean current-voltage relationship for subunit combinations as in (A); n = 29–32. Error bars indicate the mean ± SE. (C) Mean normalized tail current (at −30 mV) versus pre-pulse potential for oocytes as in (A); n = 29–32. Error bars indicate the mean ± SE. (D) Mean activation rate reported as τ of a single-exponential fit at −40 mV for the traces in (A); n = 29–32. Error bars indicate the mean ± SE; ns = p > 0.05. (E) Representative traces from TEVC recordings of Xenopus laevis oocytes injected with cRNA encoding KCNQ2/KCNQ3, alone or co-injected with cRNA encoding SMIT1; n = 10–13. (F) Mean current-voltage relationship for subunit combinations as in (E); n = 10–13. Error bars indicate the mean ± SE. (G) Mean normalized tail current (at −30 mV) versus pre-pulse potential for oocytes as in (D); n = 10–13. Error bars indicate the mean ± SE. (H) Mean activation rate reported as τ of a single exponential fit at −40 mV for traces in (E); n = 10–13. Error bars indicate the mean ± SE; ∗∗p < 0.01.