Figure 7. NMR structure of the full-length agnoprotein.
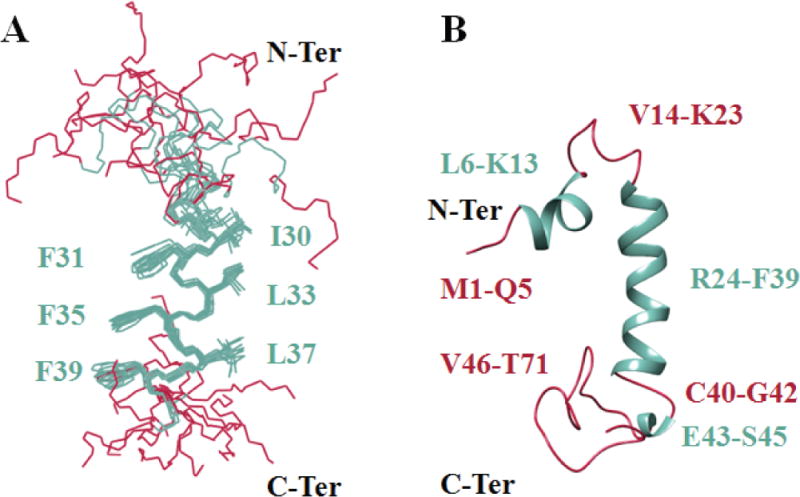
Structure was determined in presence of 30% (v/v) TFE at pH 3.0 and 313°K, using CNS [Brunger et al., 1998] and ARIA [Nilges, 1995] programs. Alpha helical regions and intrinsically unstructured regions are colored in green and red respectively. (A) Superimposition of the 12 energetically most favorable structures of the protein on the backbone atoms of domain Lys24-Phe39 of the average structure. The backbone bonds were drawn with lines. The well-defined sidechains (Phe31, Phe35, Phe39, Ile30, Leu33 and Leu37) were displayed in the stick representation. (B) Average structure of agnoprotein contains two α-helical structures Leu6- Lsy13 and Arg24-Phe39 (colored green) and two principal unstructured regions spanning residues Val14-Lys23 and Cys40-Thr71 (colored red). In some structures, a short α-helix region, Glu43-Ser45, is also formed.
