Fig. 6.
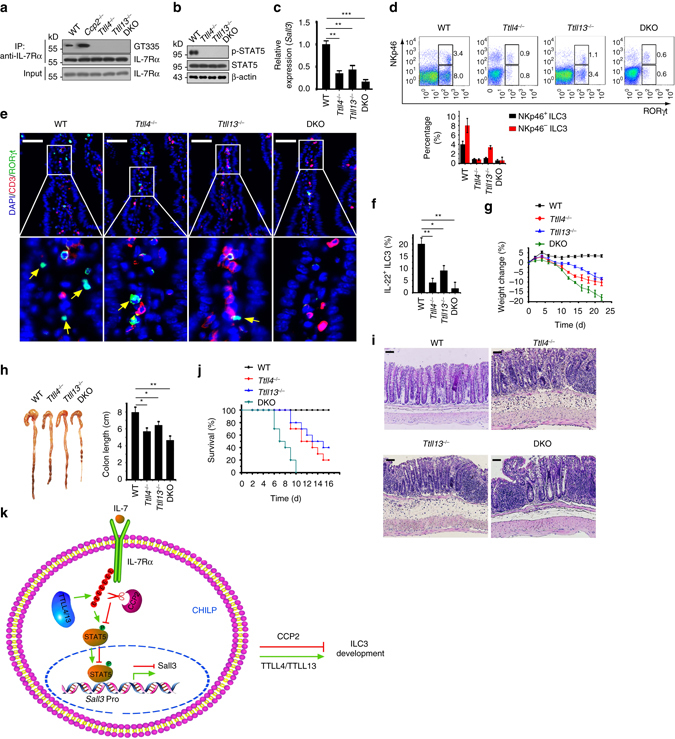
Deletion of Ttll4 or Ttll13 impairs ILC3 development. a Detection of IL-7Rα glutamylation in CHILPs by gating on Lin−IL-7Rα+Flt3−CD25−α4β7 + from WT, Ccp2 −/−, Ttll4 −/−, Ttll13 −/−, and Ttll4 −/− Ttll13 −/− mice. b Examination of STAT5 phosphorylation in CHILPs from WT, Ccp2 −/−, Ttll4 −/−, Ttll13 −/−. and Ttll4 −/− Ttll13 −/− mice. c Sall3 expression levels were tested in WT, Ttll4 −/−, Ttll13 −/−, and Ttl4 −/− Ttll13 −/− CHILPs by quantitative reverse transcription PCR (RT-qPCR). Relative fold change of Sall3 expression values were normalized to endogenous Actb. d Flow cytometry analysis of ILC3s in small intestine lamina propria from WT, Ttll4 −/−, Ttll13 −/−, and Ttll4 −/− Ttll13 −/− mice. Percentages of indicated cells were calculated and shown as means±s.d. (right panel). n = 6 for each group. e Analysis of ILC3s in WT, Ttll4 −/−, Ttll13 −/−, and Ttl4 −/− Ttll13 −/− small intestines by immunofluorescence staining in situ. Arrowhead denotes ILC3 cells. Scale bars, 50 μm. f Analysis of IL-22+ ILC3s after IL23 stimulation by flow cytometry. n = 6 for each group. g Body weight changes of WT, Ttll4 −/−, Ttll13 −/−, and Ttll4 −/− Ttll13 −/− mice post C. rodentium infection. n = 10 for each group. h Colon length from WT, Ttll4 −/−, Ttll13 −/−, and Ttll4 −/− Ttll13 −/− mice after C. rodentium infection. n = 6 per genotype group. i Histology of colons from WT, Ttll4 −/−, Ttll13 −/−, and Ttll4 −/− Ttll13 −/− mice 8 days after infection with C. rodentium. Scale bars, 50 μm. j Survival curves of WT, Ttll4 −/−, Ttll13 −/−, and Ttll4 −/− Ttll13 −/− mice after C. rodentium infection. n = 10 for each group. In all, 2 × 109 C. rodentium was used by oral inoculation. k A working model represents glutamylation-mediated IL-7 signaling in the regulation of ILC3 development. Pro promoter. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t-test). Data represent three independent experiments. Error bars in c, d, and f–h indicate s.d.
