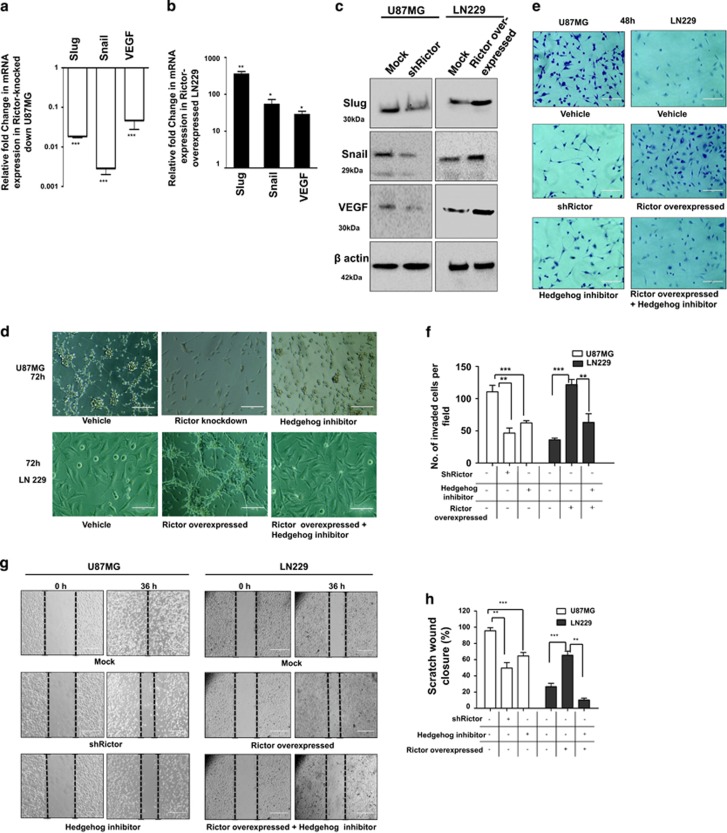
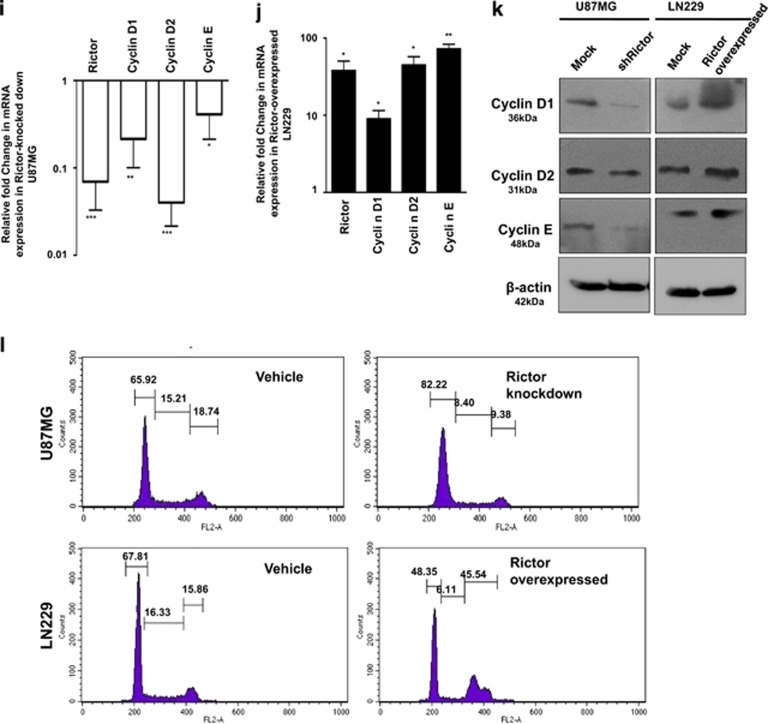
Figure 3.
mTORC2 regulates angiogenesis, invasion, migration and proliferation of cancer cells via the hedgehog pathway. U87MG cells were treated with either shRictor_1 and shRictor_2 or GANT61 (Gli2 inhibitor) (100 nM) for 24 h. Rictor was overexpressed in LN229 cells and treated similarly with GANT61 to perform the following experiments (A–L). (a) Real-time PCR analysis of Slug, Snail and VEGF mRNA expression in U87MG cells after mTORC2 disruption relative to that of shRictor-untransfected cells. Values are normalized against 18S rRNA expression (n=3 experiments). (b) Real-time PCR analysis of Slug, Snail and VEGF mRNA expression in LN229 cells after mTORC2 activation relative to that of untransfected cells. Values are normalized against 18S rRNA expression (n=3 experiments). (c) Representative immunoblots showing decreased Slug, Snail and VEGF protein levels upon reduced mTORC2 activity in U87MG cells, whereas all these protein levels were enhanced when mTORC2 activity was increased in LN229 cells. (d) Inverted light microscopic images showing reduced connective tubes formation between cellular colonies in Rictor-knocked-down and GANT61-treated U87MG cells compared with vehicle. Rictor-overexpressed LN229 cells exhibited increased connective tubes formation compared with untransfected cells, which was reduced in the presence of hedgehog inhibitor. (e and f) Treated or untreated U87MG or LN229 cells (5 × 104) were suspended in medium without FBS (100 μl) and added to the upper chamber of an insert (6.5 mm diameter, 8 μm pore size; Becton Dickson). The insert was placed in a 24-well plate containing medium (700 μl) with or without 10% FBS. Inverted light microscopic images of U87MG cells showing lower invasiveness when Rictor was knocked down or Gli2 activity was inhibited by GANT61. In contrast, LN229 cells showed increased invasion when Rictor was overexpressed. However, Rictor-overexpressed cells showed lower invasion in the presence of Gli2 inhibitor (e). Three randomly selected fields on the lower side of the insert were counted and graphically represented (n=3) (f). (g and h) U87MG and LN229 cells were cultured to >80% confluency in a six-well plate and treated with either shRictor or GANT61 for 24 h. Rictor-overexpressed LN229 cells were treated with GANT61. In every treated or untreated well, scratch was made by 2.5 μl tips. Representative images showing the filling of gaps in each well after 36 h (g). Area of closure was calculated and graphically represented (n=3) (h). (i and j) Real-time PCR analysis of Rictor, cyclin D1, cyclin D2 and cyclin E mRNA expression in Rictor-knocked-down U87MG (i) and Rictor overexpressed LN229 cells (j) relative to that of untransfected cells. Values are normalized against 18S rRNA expression (n=3 experiments). (k) Representative immunoblots showed decreased proteins level of cyclin D1, cyclin D2 and cyclin E upon reduced mTORC2 activity in U87MG cells, whereas all these protein levels were enhanced upon increased mTORC2 activity in LN229 cells. (l) Flow cytometric analysis exhibited cell cycle arrest in Rictor-knocked-down U87MG cells and enhanced proliferation in Rictor-overexpressed LN229. y axis of the graphs is denoting the cell counts and x axis is denoting the DNA area. Cycle Test Plus kit (BD Bioscience) was used for cell cycle analysis. At least 20 000 cells were acquired in FACS and analyzed by CellQuest Pro software (BD FACSCalibur). The data are presented as means± SD of three independent experiments. Statistical significance compared with the control is indicated by *P<0.05, **P<0.01 and ***P<0.001