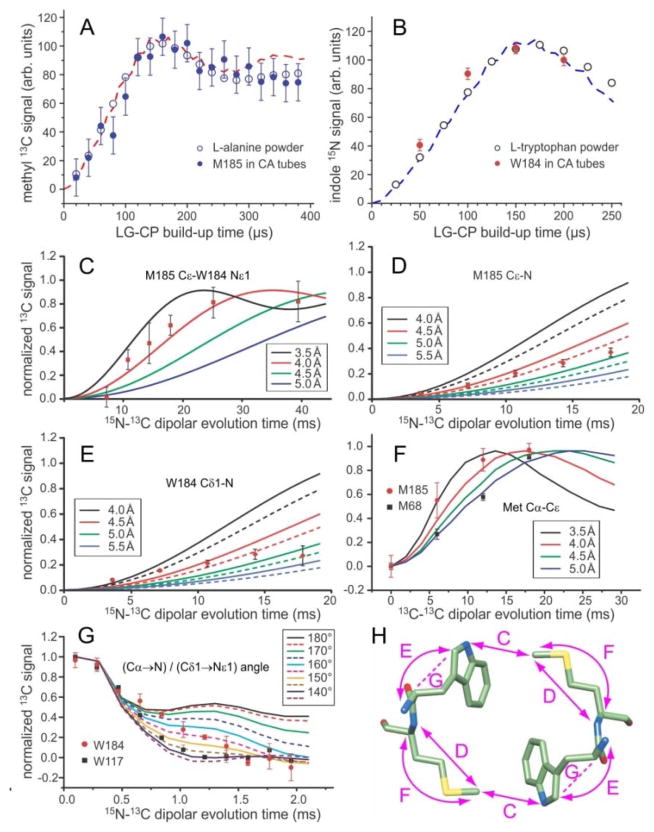
Figure 3.
Quantitative restraints on the dimerization interface structure from solid state NMR. (A,B) Comparison of 1H-13C and 1H-15N LG-CP build-up data for M185 Cε and W184 Nε1 in HIV-1 CA tubes with data for the methyl site in L-alanine powder and the Nε1 site in L-tryptophan powder. The similarity of these data for CA tubes and amino acid powders indicates the absence of large-amplitude motions of M185 and W184 sidechains on sub-millisecond timescales. Dashed lines are ideal LG-CP simulations for four-spin (panel A) and two-spin (panel B) systems, assuming methyl C-H bond lengths of 1.08 Å, with rapid methyl rotation, and indole N-H bond lengths of 0.98 Å. (C,D,E) 15N-13C REDOR data that restrain the intermolecular M185 Cε-W184 Nε1 distance, the intra-residue M185 Cε-N distance, and the intra-residue W184 Cε1-N distance in HIV-1 CA tubes. Solid curves in panel C are ideal two-spin simulations for isolated 15N-13C pairs with the indicated distances. Solid and dashed curves in panels D and E are three-spin simulations, including an additional 15N at the farthest and closest possible distance (see text). (F) BroBaRR data that restrain the intra-residue Cα-Cε distance in M185. Data for M68 are also shown for comparison. Solid lines are two-spin simulations for the indicated 13C-13C distances. (G) NCCN data that restrain the angle between the Cα-N and Cδ1-Nε1 bond vectors in W184. Data for W184 are also shown for comparison. Solid and dashed lines are ideal simulations for the indicated angles. (H) Dimer of W184-M185 units, showing the distances (double-headed arrows) and vector angles (dashed lines) restrained by data in panels C-G. Error bars in panels A–G represent uncertainties calculated from the root-mean-squared noise in the corresponding solid state NMR spectra.