Fig. 1.
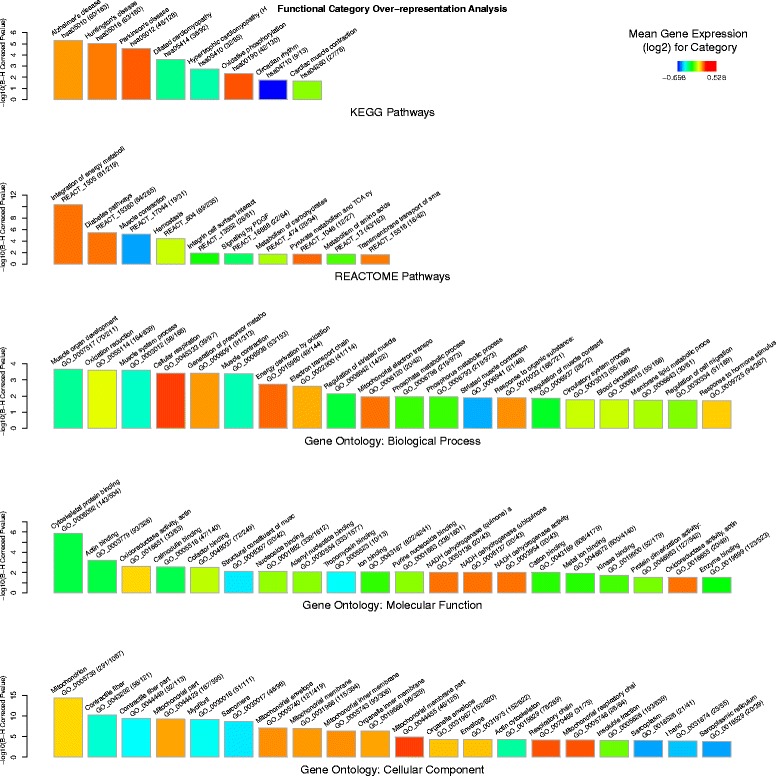
Cellular functions of the exercise response. Bar charts showing over-representation of functional categories (described by KEGG and REACTOME pathways and Gene Ontology: Biological Process, Molecular Function and Cellular Component) for the list of genes (n = 3241) that showed statistically significant differential expression, of at least +/− 1.25-fold, between the muscle tissue (gluteus medius) from the untrained rest (UR) and the untrained exercise (UE) Thoroughbred cohorts. Bars represent the most significant functional modules (up to 20) for each of the five annotation schemas. Bar height represents statistical significance (−log 10 transformed Benjamini-Hochberg (B-H) Corrected P-value) of the over-representation, based on the EASE-score (conservative Fisher Exact t-test). Bar color represents the mean differential expression (log2(UE/UR) for the genes in this module (see color key). Category name, ID and size (number category genes in gene list/category size) are given above each bar. For example it can be seen that the fourth most significant Gene Ontology:Biological Process is ‘Cellular respiration’ and that it is one of the most up-regulated functional categories on average (red color) and that 39 out of the 97 genes assigned to this category are differentially expressed between UE and UR cohorts. Full results are provided in Additional file 2: Table S3
