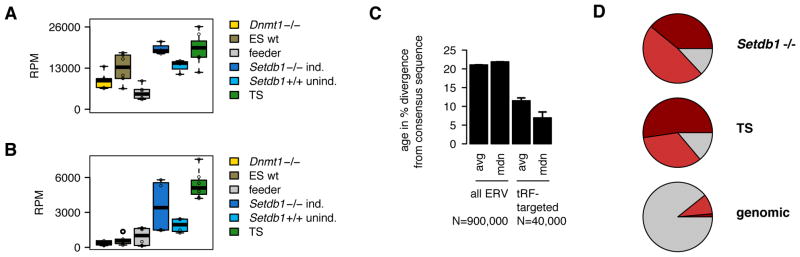
Figure 1. Stem cells with relaxed epigenetic control of LTR-retrotransposons express sRNA targeting ERVs, including 3′ tRNA-derived fragments (tRFs).
(A) Setdb1 knockout mouse ES cells and TS cells have elevated levels of LTR-retrotransposon (ERV) small RNA and (B) 3′ CCA tRFs which target ERVs. Boxplots represent reads per million total mapped reads (RPM) of biological replicates; for a list of all 33 sRNA libraries refer to Figure S3. Setdb1 knockout induced −/−, uninduced +/+; for details see STAR Methods section. (C) tRF-targeted ERVs (in TS cells) are potentially active and younger than the average genomic ERV copy (avg: average; mdn: median; ± SD). (D) The majority of ERV-K targeted by 3′ tRFs are from the ETn and IAP families which are the most active LTR-retrotransposons in mouse (average RPM values of 4 replicate Setdb1 −/− and 7 replicate TS sRNA libraries). For comparison, relative abundance of ERV-K sequences in the mm9 mouse genome: 9% belong to the IAP family, 1% to the ETn family. See also Figure S1.