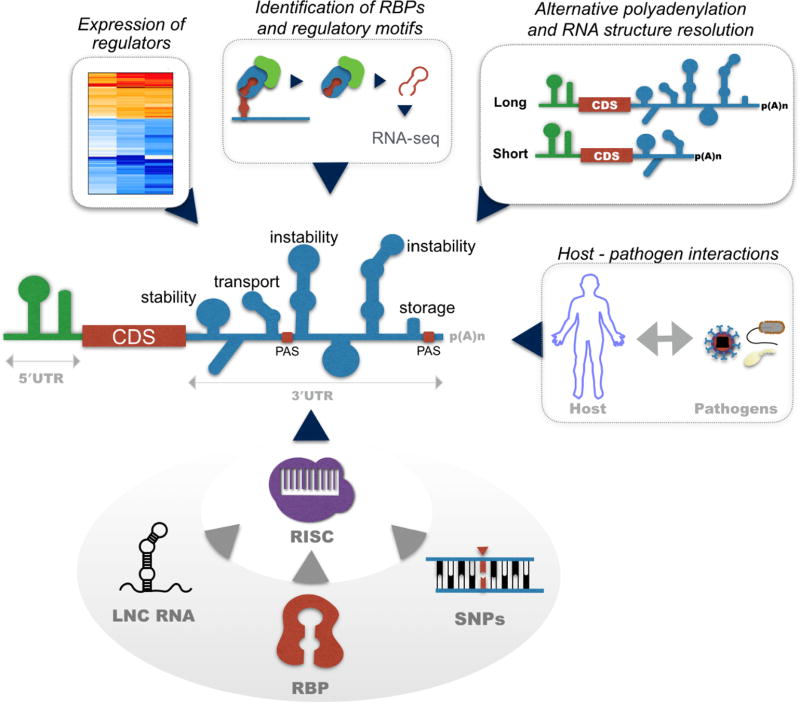
Figure 1. Schematic of microRNA interactions with post-transcriptional regulators.
Recruitment of the RNA-induced silencing complex (RISC) to miRNA recognition elements (MRE) leads to ribonuclease-mediated mRNA decay and alterations in gene expression. The co-expression of post-transcriptional modulators such as RNA-binding proteins (RBP) and long non-coding RNAs (lncRNAs) can impact miRNA biogenesis and function. RBPs can inhibit miRNA expression and prevent the miRNA recruitment to target mRNAs through competitive binding or mRNA compartmentalization. On the other hand, RBPs can cooperatively enhance miRNA-mediated mRNA decay. LncRNAs can interact with DNA, RNA and RBPs, suppressing both miRNA and RBP activity through competitive binding. Additionally, the function of miRNAs can be impacted by changes in the sequence and structure of the 3′ UTR. Single nucleotide polymorphisms (SNPs) can disrupt or create novel MREs. SNPs can also change the structure of the 3′ UTR, hindering the accessibility of miRNA and RBPs to their target elements. Finally, dynamic changes in the length of the 3′ UTR through usage of alternative polyadenylation sites (pA) results in changes in the number of potential interaction sites/motifs for the previously mentioned post-transcriptional modulators, impacting mRNA stability. The development of novel molecular and computational tools provide means to understand global changes in post-transcriptional regulator expression, the identification of mRNA regulatory motifs, and changes in the sequence and structure of the 3′ UTR. Building a comprehensive post-transcriptional regulome will allow us to contextualize the function of miRNAs in the regulation of host gene expression during immune development, infection, and immunological disease.