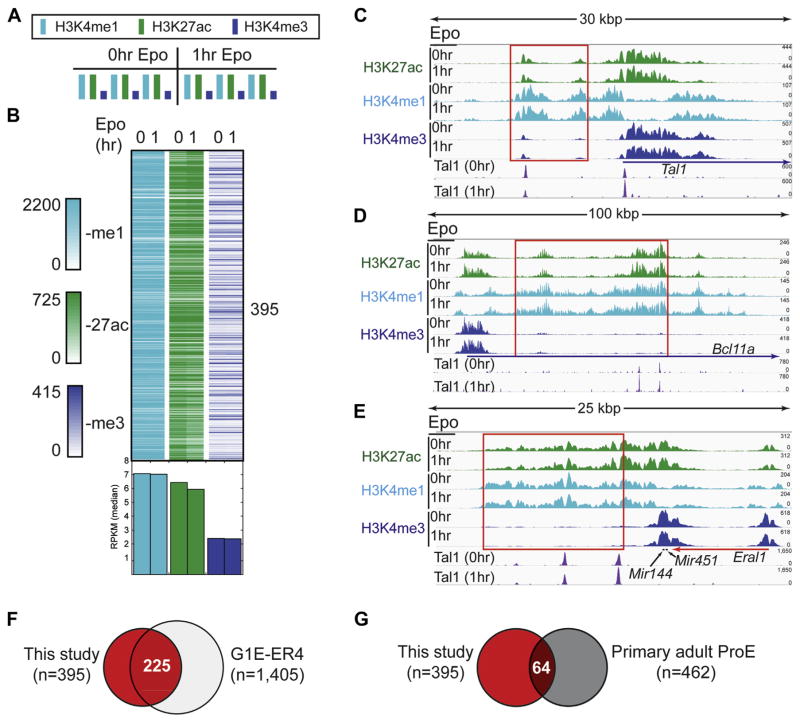
Figure 6.
Identification of super-enhancers at erythroid genes involved in cell fate decisions. (A) Histone modification enrichment across super-enhancers in erythroid cells before and after Epo stimulation. (B) Density plots illustrating the H3K4me1, H3K27ac, and H3K4me3 ChIP-exo signal (RPKM normalized) across super-enhancer intervals before and after Epo treatment. The normalized RPKM median value for each column is represented as a bar graph below the density plots. (C–E) Genome browser views of super-enhancers before and after Epo stimulation at the Tal1, Bcl11a, and Mir144/Mir451 genes, respectively. Red boxes denote genomic intervals identified as super-enhancers by the Whyte et al. algorithm employed by HOMER. (F,G) Venn overlap of super-enhancer intervals identified in this study (red fill) compared with super-enhancer regions from Huang et al. [18] in mouse (panel F, G1E-ER4) and human (panel G, primary adult ProE) erythroid cells, respectively.