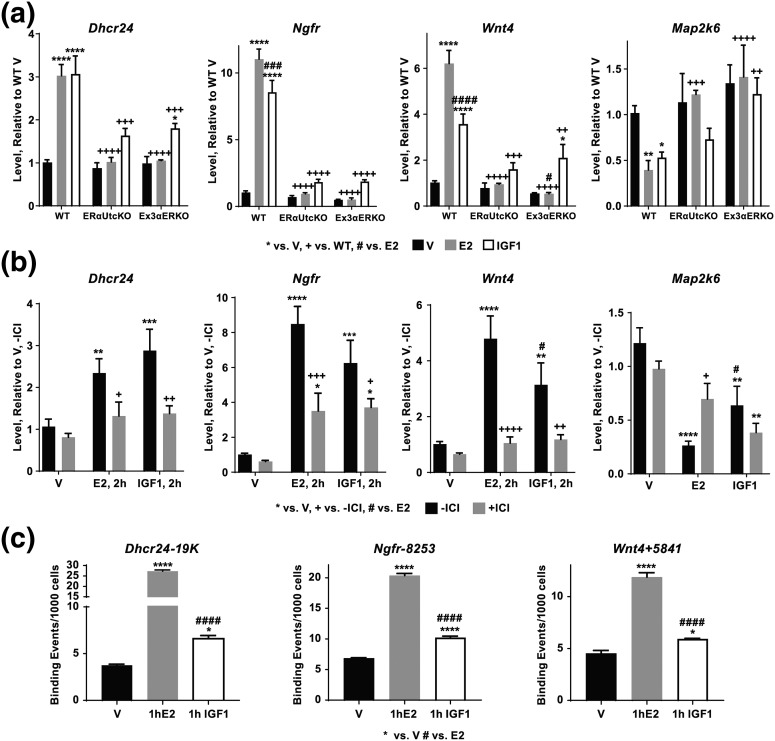
Figure 5.
Verification of ERα-dependent transcriptional responses. (a) RT-PCR showing E2 (2 hours) and IGF1 (2 hours) induction of Dhcr24, Wnt4, and Ngfr and repression of Map2k6 is ERα dependent. Two-way analysis of variance with posttest *P < 0.05, **P < 0.01, ***P < 0.001 vs V; ++P < 0.01, +++P < 0.001, ++++P <0.0001 vs WT; ###P < 0.001, ####P < 0.0001 E2 vs IGF1. N = 3 to 7 for all samples. Black = V, gray = E2, white = IGF1. (b) ICI inhibits E2 (2 hours) and IGF1 (2 hours) induction of Dhcr24, Wnt4 and Ngfr, consistent with a two-way analysis of variance with posttest *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 vs V; +P < 0.05, +++P < 0.001 vs –ICI; #P < 0.05 E2 vs IGF1. N = 4 to 5 for all samples. Black = no ICI, gray = with ICI. (c) ChIP-PCR confirms binding of ERα to sites near the Dhcr24, Ngfr, and Wnt4 transcripts. All three tested sites have basal ERα binding (V) that is increased by E2 or IGF1 activation. *P < 0.05, ****P < 0.0001 vs V; #### P < 0.0001 E2 vs IGF1. Black = V, gray = E2, white = IGF1. A pool of three uterine samples was tested per treatment; assay done in triplicate.