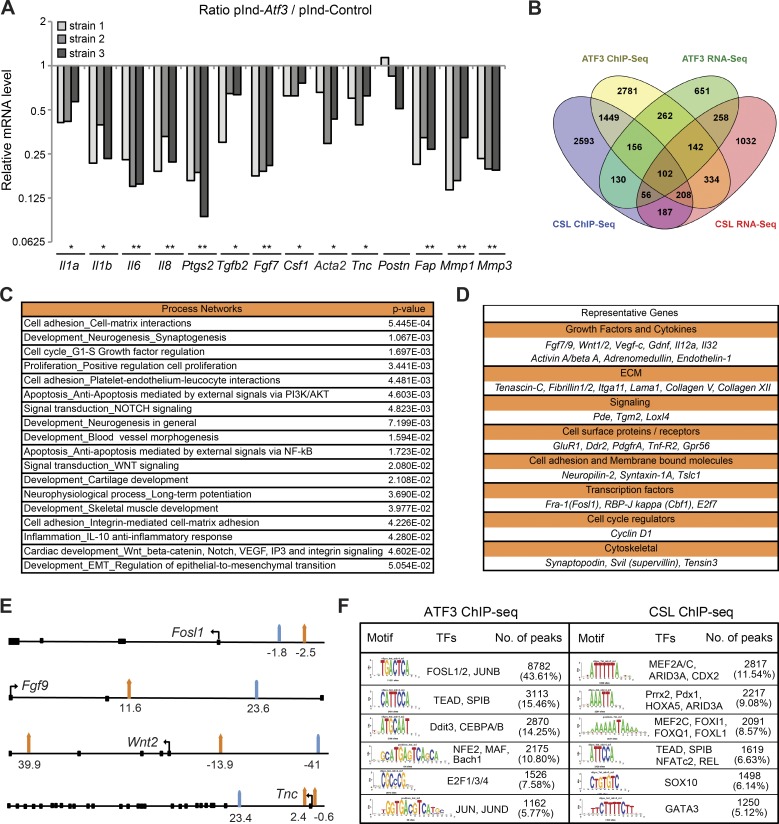
Figure 3.
Induced Atf3 suppresses CAF effector genes with large overlap of binding targets with CSL. (A) Several HDF strains infected with a lentiviral vector for Doxycycline (Dox)–inducible Atf3 expression (pInd-Atf3; n = 3) versus empty vector control (pInd-Ctr; n = 2) were analyzed with and without Dox treatment (750 ng/ml for 3 d) for expression of the indicated genes. Values are expressed as ratio log10 (pInd-Atf3/pInd-Ctr), two-tailed, one-sample t test, *, P < 0.05, **, P < 0.005. (B) Overlap between ChIP-seq and RNA-seq profiles of genes bound and modulated by increased ATF3 expression in HDFs and those previously identified to be bound and controlled by CSL (Procopio et al., 2015). A complete list of genes is provided in Table S1. (C) GeneGo MetaCore analysis was performed with the data set of common direct target genes regulated by induced ATF3 and CSL. Shown is a list of process networks with statistically significant enrichment (P < 0.05). The complete list is provided in Table S2. (D) Representative list of common directly regulated genes by ATF3 and CSL divided by biological function. (E) Schematic representation of induced ATF3 (orange arrows) and CSL (turquoise arrows) binding peak positions for four genes commonly targeted and regulated by induced ATF3 and CSL listed in D. Coding exons are shown as black squares, and the directions of the transcription start sites are shown as black arrows. A complete list of induced ATF3 and CSL binding peak positions is provided in Table S3. (F) Motif enrichment analysis of nucleotide sequences surrounding elevated ATF3 and CSL binding peaks was performed by the RSAT peak-motifs tool. The number of ATF3 or CSL binding peaks with at least one predicted site of the indicated transcription factors is indicated along with the corresponding frequency (percentage).