Fig. 1.
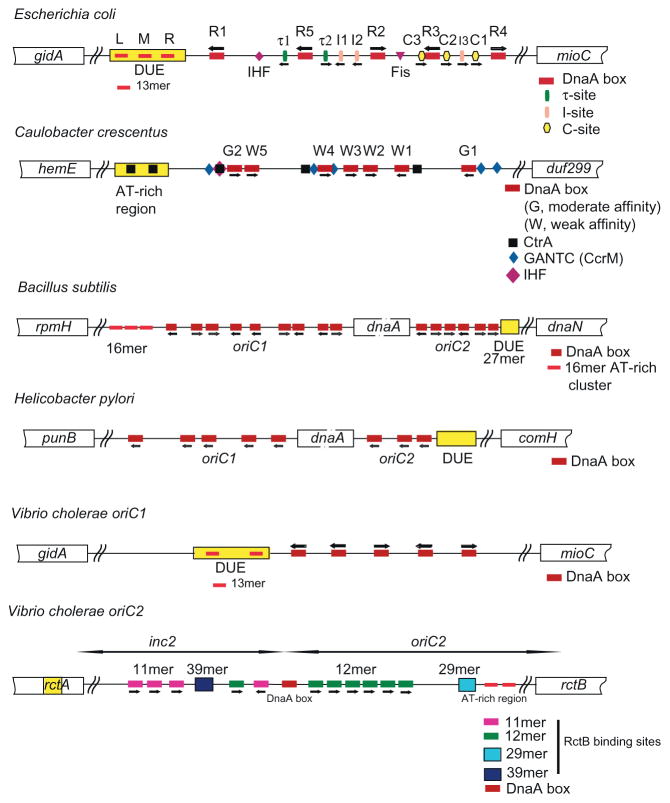
Bacterial replication origins carry DNA sequence motifs recognized by DnaA and other proteins, and an AT-rich region named the DNA unwinding element (DUE). Among bacteria however, the presence, relative arrangement, and position of these DNA motifs are not conserved. For E. coli, DnaA in a complex with either ATP or ADP recognizes a specific DnaA box with comparable affinity, but the relative affinity among the individual DnaA boxes varies. In comparison, DnaA-ATP and not DnaA-ADP specifically binds to I-, τ-, and C-sites. The affinities of DnaA-ATP to these sites are less than to the DnaA boxes. E. coli oriC also carries binding sites for Fis and IHF, whereas the replication origin of C. crescentus contains a binding site for IHF but not Fis. In contrast with these replication origins, which are contiguous DNA sequences, the replication origins of B. subtilis and H. pylori are bipartite in which oriC1 and oriC2 are separated by the dnaA gene. B. subtilis oriC1 has three AT-rich 16mer repeats upstream of dnaA [34,35]. A 27mer AT-rich cluster in the oriC2 region is unwound by DnaA [36]. V. cholerae carries two chromosomes. Initiation from oriC1 appears to be similar to E. coli oriC. Initiation from oriC2 requires RctB, which recognizes 11mer and 12mer iterons, and also a 39mer and a truncated form of the 39mer named the 29mer. The 39mer in the inc2 region and the 29mer bound by the monomeric form of RctB together with the DnaA box bound by V. cholerae DnaA regulate the frequency of initiation from oriC2 [37]. Adapted from Refs. [36–38].