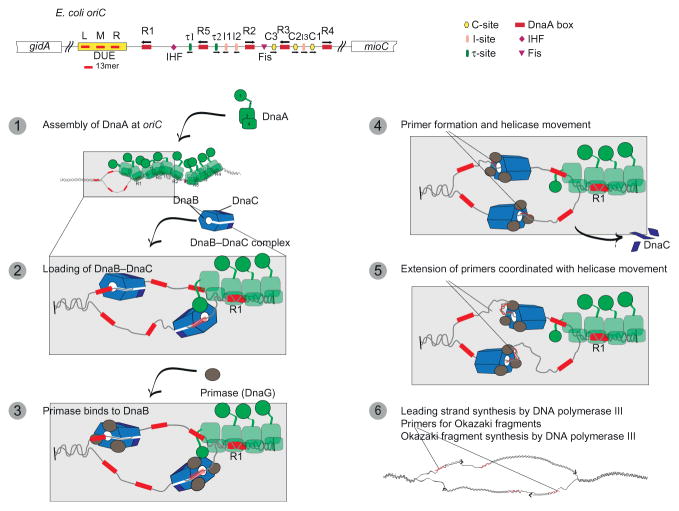
Fig. 3.
Assembly of DnaA, and the DnaB–DnaC complex at E. coli oriC, and activation of DnaB is a step-wise process. Step 1: DnaA recognizes specific DNA elements in E. coli oriC (see Fig. 1 and the text) to assemble a DnaA oligomer at this site. The numbers in the figure of DnaA represent its functional domains. Upon assembly of DnaA-ATP as a self-oligomer, it induces unwinding of a region containing the 13mers by binding to the top strand. At this step, domain IV of DnaA binds to the DnaA boxes and presumably to C-, I-, and τ-sites whereas domain I and III of a DnaA monomer interacts with another to assemble the DnaA oligomer. The interaction of HU with domain I of DnaA stabilizes the DnaA oligomer, but this interaction is not shown for simplicity. Step 2: Domain I of DnaA interacts with the N-terminal domain of DnaB in the DnaB6–DnaC3 complex to load this complex onto each DNA strand of the unwound region. Step 3: Primase interacts with the N-terminal domain of DnaB, which is required for primer synthesis. Step 4: As primase synthesizes a primer (shown as red (dark gray in the print version) squiggles) on the top and bottom strands, this event is coupled with the translocation of DnaB and the dissociation of DnaC from DnaB. Step 5: Primase completes primer synthesis and will then dissociate from DnaB after the primer is transferred to DNA poly-merase III holoenzyme. DnaB moves on each single-stranded DNA toward the junction of each replication fork. Step 6: DNA polymerase III holoenzyme (not shown) extends these primers for the synthesis of each leading strand. During this elongation stage of DNA replication, DnaB at the junction of each replication fork will act as a DNA helicase to unwind the parental duplex DNA. The translocation of DnaB and its transient interaction with primase leads to the synthesis of subsequent primers on the top and bottom DNA strands. These primers are used by DNA polymerase III holoenzyme for the synthesis of Okazaki fragments.