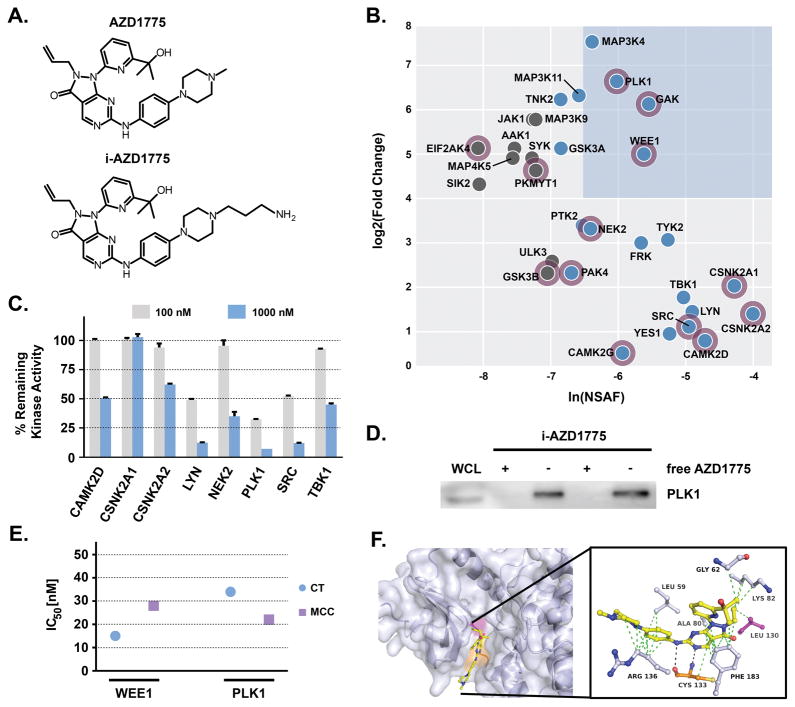
Figure 2. Target profile of AZD1775 in H322 NSCLC cells.
(A) Chemical structures of AZD1775 and its immobilizable analogue i-AZD1775. (B) Protein kinase interaction profile of AZD1775 in H322 cells. Blue box highlights prominent kinase target candidates with high spectral abundance (based on NSAF: normalized spectral abundance factor) and specificity (based on fold change of spectral counts between i-AZD1775 affinity pulldowns and competition experiments with unmodified AZD1775). Blue nodes indicate kinases with high NSAF (> −7), grey nodes with low NSAF (< −7). Purple halo indicates kinases with gene ontology annotation for cell cycle. (C) Inhibition of identified kinases by 100 nM or 1000 nM AZD1775 in in vitro kinase assays (Reaction Biology). (D) PLK1 immunoblot of i-AZD1775 affinity pulldowns (−) and AZD1775 competition pulldowns (+) from H322 whole cell lysates (WCL). (E) Comparison of WEE1 and PLK1 inhibition by AZD1775 in in vitro kinase assays (Eurofins). Displayed are the IC50 values observed for both kinases using commercial (CT: Chemietek) and in-house synthesized (MCC: Moffitt Cancer Center) AZD1775. (F) In silico docking of AZD1775 to PLK1 based on the X-ray co-crystal structure of PLK1 with BI-2536, PDB: 2RKU.