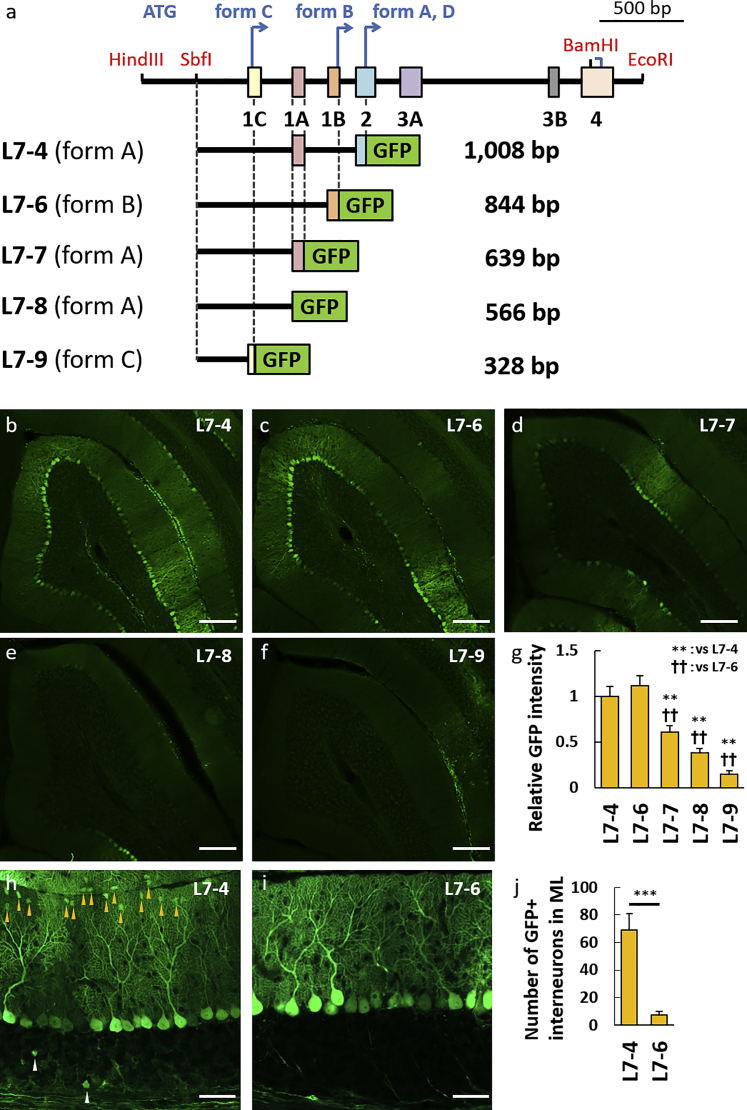
Figure 4.
The 0.8-kb Genome Region Upstream of the Transcription Initiation Codon in Exon 1B, L7-6, Shows a Similar Promoter Strength as L7-4 and Much Less Leaky Activity in Cortical Cells Other Than Purkinje Cells
(A) Schema depicting the L7-4 promoter and the 5′ side deletion constructs. Whereas the L7-4 promoter uses the transcription initiation codon in exon 2 as the form A and form D, the L7-6 and L7-9 promoters use different transcription initiation codons in exon 1B (L7-6) and exon 1C (L7-9) as the form B and form C, respectively. The L7-7 and L7-8 promoters are present or absent from exon 1A in the form A, respectively. The size (base pairs [bp]) of each construct without the GFP sequence is shown at the right side of each scheme. (B–F) Representative native GFP fluorescent images on sagittal sections of the cerebellum virally expressing GFP by the deleted promoters as depicted. Robust GFP expression was observed by L7-4 (B) and L7-6 (C), whereas the GFP expression was significantly decreased by deletion of the 3′ side (D–F). (G) Summary graph showing the GFP intensity relative to that by the L7-4 promoter. F(4,39) = 19.96; p < 0.001, ANOVA; **p < 0.01; ††p < 0.01 by Bonferroni correction for post hoc test, as compared with L7-4 or L7-6 promoter, respectively. Error bars indicate SEM. (H and I) Enlarged GFP fluorescent images of the cerebellar cortex virally expressing GFP by the L7-4 (H) or L7-6 (I) promoter. Arrowheads indicate GFP-labeled non-Purkinje cells. (J) Graph showing the number of GFP-positive non-Purkinje cells in the molecular layer. t(10) = 4.86; ***p = 0.001 by Student’s t test. Error bars indicate SEM. Statistical data were obtained using 11 (L7-4), 12 (L7-6), 8 (L7-7), 8 (L7-8), and 5 (L7-9) mice. Scale bars, 200 μm (B–F) and 50 μm (H and I).