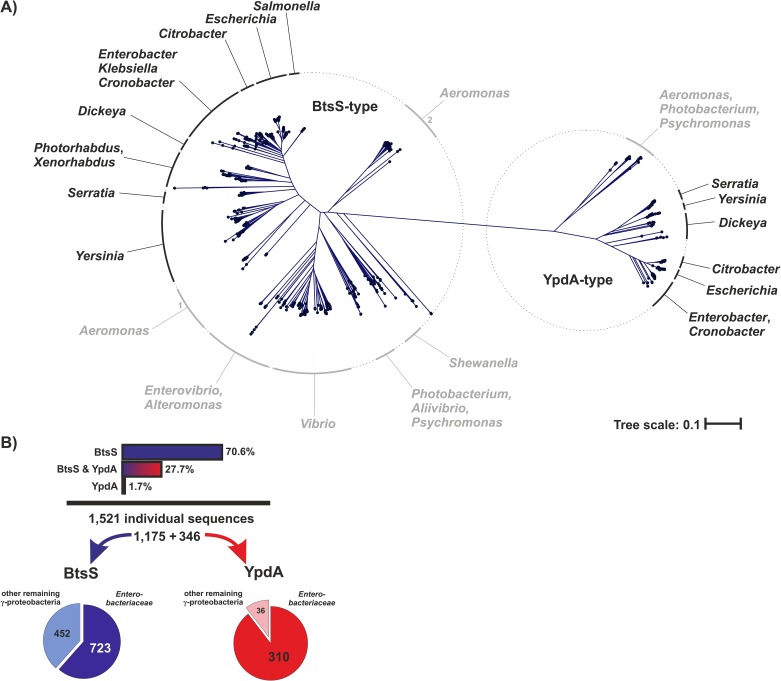
Fig 2. Phylogenetic tree of LytS-type histidine kinases in γ-proteobacteria.
(A) Based on a local alignment search using the NCBI RefSeq protein database, 1,521 individual LytS-type sequences were identified, and amino acid identity was calculated using the progressive alignment algorithm of the CLC Main Workbench 7.6 software. The phylogenetic tree was created using CLC’s high-accuracy neighbor-joining algorithm with 100 bootstrap replicates. Reassigned genera are given in black for the predominant members of the Enterobacteriaceae and in grey in the case of other γ-proteobacterial families. All genes encoding BtsS- or YpdA-type proteins in the set of species included in the tree are listed in S1 Table. The lengths of the branches of the tree represent the relative amount of evolutionary divergence between any two sequences in the tree. (B) Summary of the relative abundance of BtsS and YpdA sequences detected, and their distribution among Enterobacteriaceae and other γ-proteobacteria based on the phylogenetic tree.