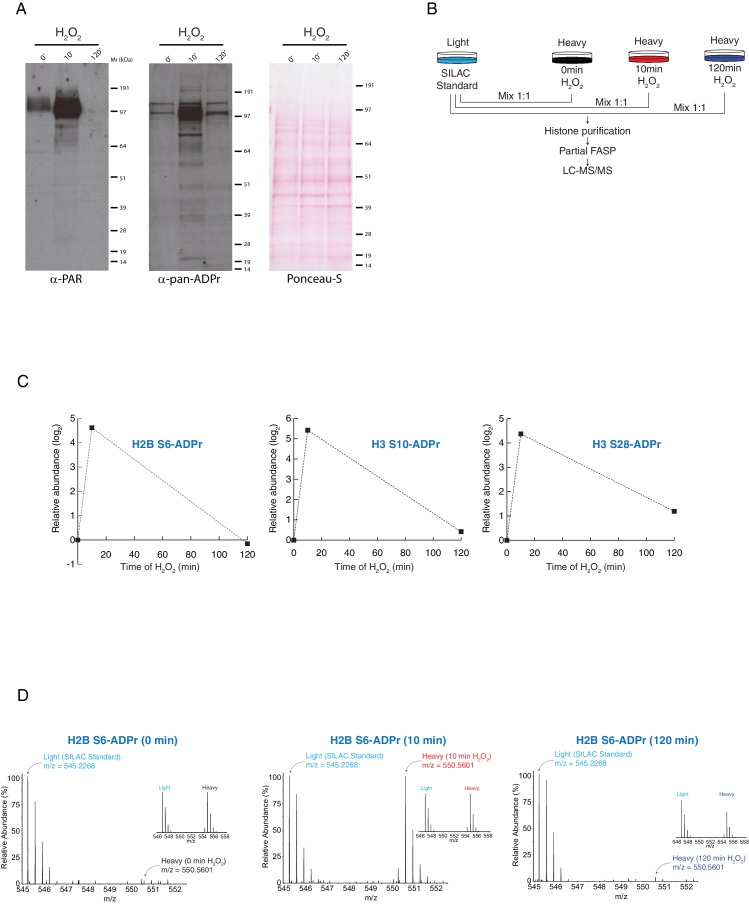
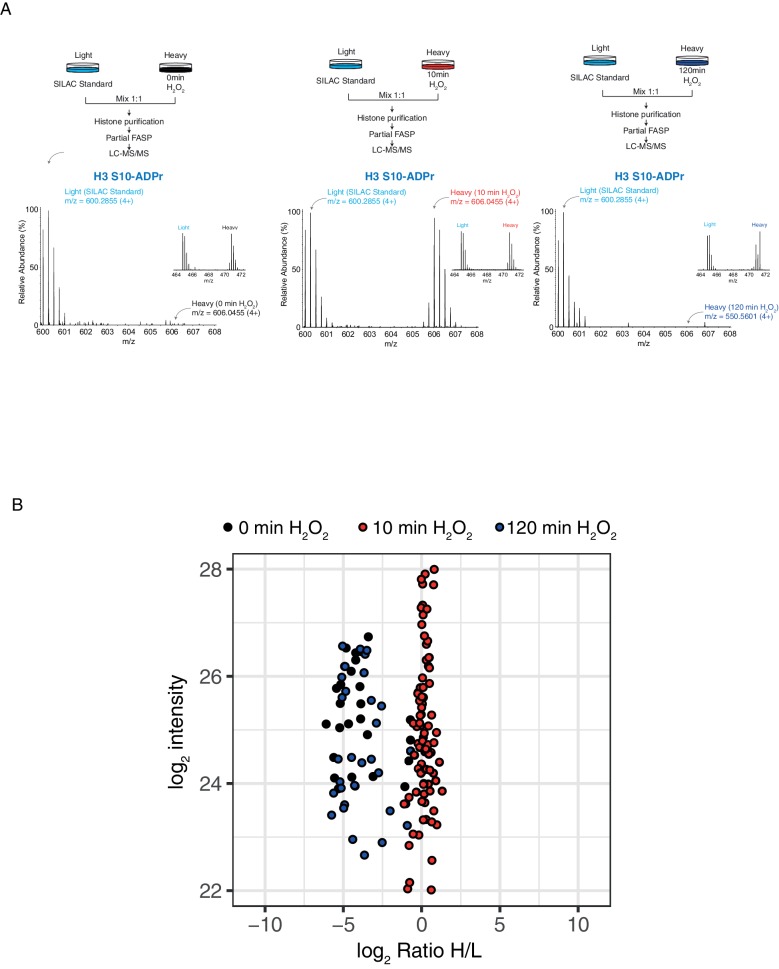
Figure 1. Histone serine ADP-ribose modification is reversible.
U2OS cells were treated with H2O2 and analysed at indicated time-points. (A) The samples were lysed and the proteins were separated by SDS-PAGE, analysed by western blot and probed for PAR (left) or pan-ADPr (middle). Ponceau-S staining was used as loading control (right). (B) Schematic representation of the SILAC-based strategy to quantify core histone Ser-ADPr marks after different time points of 2 mM H2O2–induced DNA damage. Light labeled (Lys0) cells treated for 10 min with 2 mM H2O2 were used as SILAC Standard. (C) The relative abundance of Ser-ADPr modification on histone proteins H2B (Ser6) and H3 (Ser10, Ser28) was calculated and plotted as a function against time of H2O2 treatment. (D) MS1s of a Ser-ADPr H2B peptide (H2B Ser6-ADPr) at different time points of 2 mM H2O2 treatment. The heavy peptide was derived from cells treated with H2O2 for the indicated time point, and the light peptide was derived from the SILAC Standard (10 min H2O2-treated cells). Each inset (right) shows a ∼1:1 ratio (heavy/light) of a non-ADP-ribosylated peptide from the same experiment.
DOI: http://dx.doi.org/10.7554/eLife.28533.003