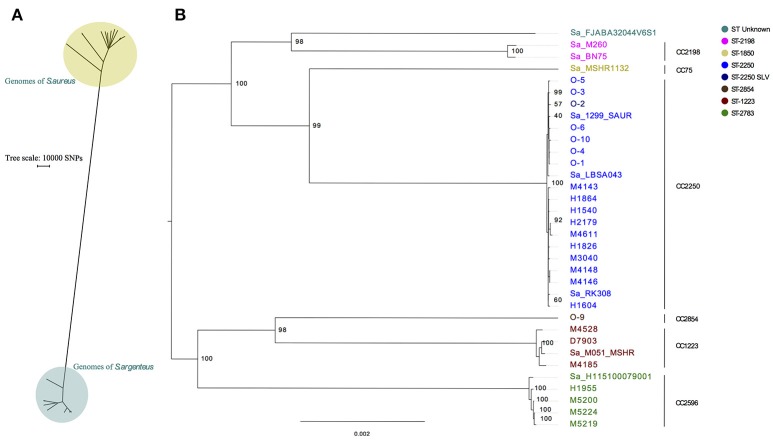
Figure 1.
Phylogenetic comparison of S. argenteus with S. aureus and of S. argenteus isolates alone. (A) SNP analysis of 34 S. argenteus genomes with a representative set of 22 genomes of S. aureus all with different sequence types. (B) Maximum likelihood phylogenetic tree of S. argenteus isolates. The tree is made from multiple sequence alignment of assemblies from 25 isolates from Denmark and 9 references of S. argenteus assemblies. One Hundred bootstraps were performed. Prefix of Sa indicates S. argenteus reference genomes downloaded from NCBI, O- prefix are Odense isolates and the remaining isolates are from Copenhagen. Bootstrap values are shown at the major breakpoints. The scale bar represents the mean number of nucleotide substitutions per site. The tree is midpoint rooted.