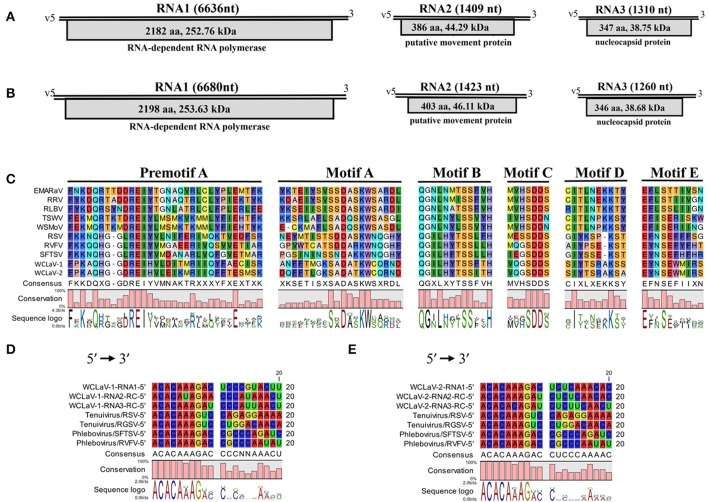
Figure 2.
Schematic representation of the genome organizations of WCLaV-1 (A) and WCLaV-2 (B). Open reading frames (ORFs) and deduced products of each RNA are shown as gray boxes, with the amino acid (aa) length, estimated molecular weight (kDa), and function of the putative proteins. (C) Amino acid alignment between conserved RdRp premotif A and motifs A–E of WCLaV-1, WCLaV-2, and selected (-ss) RNA viruses. The conserved nucleotide stretches are present at the termini of WCLaV-1 (D) or WCLaV-2 (E) with tenuiviruses and phleboviruses. RC, reverse and complementary sequence; nt, nucleotides; v, viral RNA; EMARaV, European mountain ash ringspot-associated virus (AY563040); RRV, rose rosette virus (HQ871942); RLBV, raspberry leaf blotch virus (FR823299); TSWV, tomato spotted wilt virus (D10066); WSMoV, watermelon silver mottle virus (AF133128); RSV, rice stripe virus (D31879); RVFV, Rift Valley fever virus (DQ375403.1); SFTSV, severe fever with thrombocytopenia syndrome (SFTS) virus (HM745930); RGSV, rice grassy stunt virus (AB009656).