Figure 8.
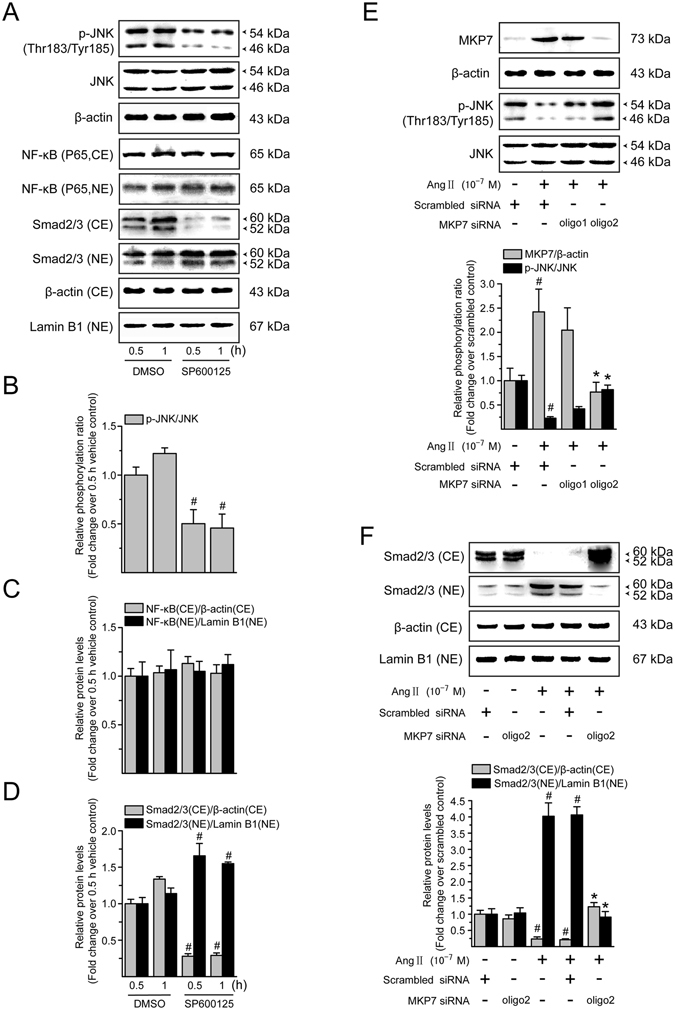
Ang II-mediated Smad activation involves JNK dephosphorylation in LX-2 cells. (A) Serum-starved LX-2 cells were incubated with SP600125 (a JNK inhibitor; 10−5 M) for 0.5 and 1 h. Whole cell extracts were immunoblotted with antibodies against JNK and its phosphorylated form. Total protein level of JNK in whole cell extracts was used as an internal control. Cytoplasmic extracts (CE) and nuclear extracts (NE) were subjected to immunoblotting with antibodies against NF-κB p65 and Smad2/3. β-Actin and lamin B1 protein levels were used as internal controls, respectively, for the CE and NE. (B) Levels of phosphorylated JNK in whole cell lysates, and (C) that of NF-κB p65 and (D) Smad2/3 in cytoplasmic and nuclear extracts, were converted to arbitrary densitometric units, normalized to the corresponding internal control and expressed relative to the phosphorylation ratio or to the protein level in cells treated with vehicle for 0.5 h (defined as 1-fold). (E) Serum-starved LX-2 cells were transiently transfected with scrambled or MKP7 siRNA. After transfection for 48 h, cells were incubated with serum free medium containing Ang II (10−7 M) for a further 0.5 h. To ensure the adequate knockdown of MKP7 and inhibition of JNK dephosphorylation, whole cell lysates were subjected to immunoblotting analysis with antibodies against MKP7 and phosphorylated JNK. The expression of β-actin and total JNK was monitored to confirm equal protein loading. (F) Cytoplasmic extracts (CE) and nuclear extracts (NE) were prepared and levels of Smad2/3 were evaluated by Western blotting. β-Actin and lamin B1 protein signals served as controls for protein loading of the CE and NE, respectively. Results of Smad2/3 protein expression in cytoplasmic and nuclear fractions were obtained from densitometric analysis, normalized by corresponding internal control and expressed relative to the protein level in scrambled siRNA-treated control cells (defined as 1-fold). All data are shown as mean ± SD of 3 independent experiments. # P < 0.05 versus vehicle- or scrambled siRNA-treated control cells; * P < 0.05 versus Ang II + scrambled siRNA-treated cells.
