Figure 1.
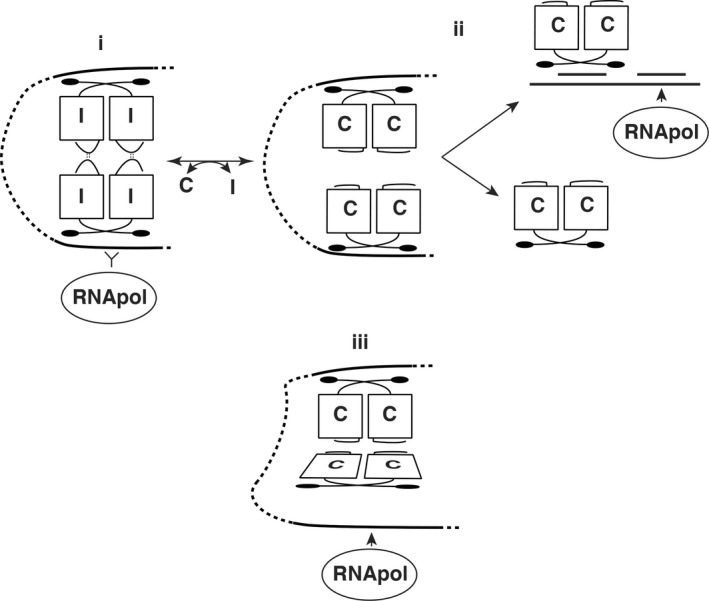
Models of PrgX function. Part i. shows a repressing complex of PrgX/I tetramers, where both XBSs are tightly bound by PrgX, preventing access RNA polymerase to the prgQ promoter. Part ii. depicts a previous working model for I is replacement by C in a tetramer, changing the C‐terminal structure of PrgX by moving a predicted tetramer‐stabilizing loop of the protein (Kozlowicz et al., 2006; Shi et al., 2005). Dissociation of the tetramer would weaken the DNA loop and favor PrgX dissociation from XBS2, allowing RNA polymerase to access the promoter. Part iii. Depicts the current working model, based on new results reported here. In this model, both peptides promote tetramer formation and looping, but the PrgX/C tetramer is distorted, placing torsional stress on the DNA loop structure reducing tight binding of PrgX to XBS2, and enabling RNA polymerase to compete more effectively for binding to the promoter. Conversion from i. to iii. occurs by replacement of one form of PrgX with another on the DNA rather than replacement of one peptide with the other in a preformed tetramer
